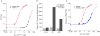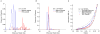Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response - PubMed (original) (raw)
Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response
Brooke M Gardner et al. Science. 2011.
Abstract
The unfolded protein response (UPR) detects the accumulation of unfolded proteins in the endoplasmic reticulum (ER) and adjusts the protein-folding capacity to the needs of the cell. Under conditions of ER stress, the transmembrane protein Ire1 oligomerizes to activate its cytoplasmic kinase and ribonuclease domains. It is unclear what feature of ER stress Ire1 detects. We found that the core ER-lumenal domain (cLD) of yeast Ire1 binds to unfolded proteins in yeast cells and to peptides primarily composed of basic and hydrophobic residues in vitro. Mutation of amino acid side chains exposed in a putative peptide-binding groove of Ire1 cLD impaired peptide binding. Peptide binding caused Ire1 cLD oligomerization in vitro, suggesting that direct binding to unfolded proteins activates the UPR.
Figures
Fig. 1
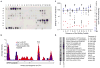
An unfolded protein co-immunoprecipitates with Ire1. A. The UPR activated by inducible CPY/CPY*-HA was measured with a 4×UPRE-GFP reporter. Fold induction is the ratio of median FITC intensity of cells in 2% galactose to cells in 2% raffinose after 4 hours. Error bars are SD; N≥4. B. FLAG-tagged Ire1 was immunoprecipitated from cells expressing CPY-HA or CPY*-HA (2 hours, 2% galactose); CPY/CPY*-HA was detected with anti-HA. The asterisk indicates unglycosylated CPY/CPY*.
Fig 2
Ire1 binds specific regions in CPY* containing basic and hydrophobic residues. A. Binding of 500 nM GST-cLD to a peptide array tiling along the sequence of CPY* was detected with anti-GST. B. The contribution of each amino acid in CPY*'s sequence to Ire1-cLD binding (WT and W426A) was calculated by averaging the intensity of spots containing that amino acid (see Methods). Values were plotted along the sequence of CPY*. SP: signal peptide. C. An array composed of peptides with single amino acid substitutions in peptide F17 from the CPY* tiling array (AQLAPYQRTGRNVYD) was probed with GST-cLDW426A and GST-cLD (Fig. S3). The binding intensity of the mutated peptides was normalized to the intensity of the wild-type peptide. Large arrows highlight the basic residues in F17. Blue: introduced basic residues. Red: introduced acidic residues. D. A peptide array of chaperone substrates shows that GST-cLDW426A and Kar2 bind a different subset of peptides.
Fig 3
A. Fluorescence anisotropy of ΔEspP-FAM (50 nM) and FAM-F17 (100 nM) binding to Ire1-cLDW426A. K1/2 for ΔEspP-FAM is 0.75±0.03 μM with a Hill coefficient of 1.2 and K1/2 for FAM-F17 is 172±4 μM with a Hill coefficient of 1.4. B. Induction of a HAC1 mRNA splicing reporter after a 3 hour incubation with 0 or 5 mM DTT in CRY1 Δ_ire1_∷KAN strains expressing no Ire1, WT Ire1, or Ire1-ΔMFY. N=5; error bars are SD; p value calculated by Student T-test between WT and ΔMFY. C. ΔMFY increases the K1/2 for Ire1-cLDW426A binding to ΔEspP-FAM to 42±2 μM, while the Hill coefficient decreases to 0.9. For A and C, N≥3; error bars are SD.
Fig 4
The addition of peptide causes Ire1 oligomerization. A. C(s) analysis of velocity sedimentation of 7 μM Ire1-cLD WT (blue) and with 50 μM ΔEspP (red). Without peptide: RMSD: 0.0105, f/f0: 1.65. With peptide: RMSD: 0.0080, f/f0: 2.75. B. C(s) analysis of the velocity sedimentation of 9 μM Ire1-cLDW426A (blue) and with 20 μM of ΔEspP (red). Without peptide: RMSD=0.0104, f/f0: 1.46. With peptide: 0.0086, f/f0: 1.52. C. Comparison of the sedimentation equilibrium (20,000×g, 12 hrs) of Ire1-cLDW426A (blue) and with a 2:1 molar ratio of ΔEspP (red) with simulated Ire1 monomer (dotted) and dimer (solid). The full data set and fit to a monomer-dimer model is presented in Fig. S8.
Comment in
- Cell biology. Sensing ER stress.
Kawaguchi S, Ng DT. Kawaguchi S, et al. Science. 2011 Sep 30;333(6051):1830-1. doi: 10.1126/science.1212840. Science. 2011. PMID: 21960615 No abstract available.
Similar articles
- On the mechanism of sensing unfolded protein in the endoplasmic reticulum.
Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. Credle JJ, et al. Proc Natl Acad Sci U S A. 2005 Dec 27;102(52):18773-84. doi: 10.1073/pnas.0509487102. Epub 2005 Dec 19. Proc Natl Acad Sci U S A. 2005. PMID: 16365312 Free PMC article. - BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response.
Pincus D, Chevalier MW, Aragón T, van Anken E, Vidal SE, El-Samad H, Walter P. Pincus D, et al. PLoS Biol. 2010 Jul 6;8(7):e1000415. doi: 10.1371/journal.pbio.1000415. PLoS Biol. 2010. PMID: 20625545 Free PMC article. - Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins.
Kimata Y, Ishiwata-Kimata Y, Ito T, Hirata A, Suzuki T, Oikawa D, Takeuchi M, Kohno K. Kimata Y, et al. J Cell Biol. 2007 Oct 8;179(1):75-86. doi: 10.1083/jcb.200704166. J Cell Biol. 2007. PMID: 17923530 Free PMC article. - How transmembrane proteins sense endoplasmic reticulum stress.
Kohno K. Kohno K. Antioxid Redox Signal. 2007 Dec;9(12):2295-303. doi: 10.1089/ars.2007.1819. Antioxid Redox Signal. 2007. PMID: 17896870 Review. - How IRE1 reacts to ER stress.
Ron D, Hubbard SR. Ron D, et al. Cell. 2008 Jan 11;132(1):24-6. doi: 10.1016/j.cell.2007.12.017. Cell. 2008. PMID: 18191217 Review.
Cited by
- MicroRNAs and obesity-induced endothelial dysfunction: key paradigms in molecular therapy.
Ait-Aissa K, Nguyen QM, Gabani M, Kassan A, Kumar S, Choi SK, Gonzalez AA, Khataei T, Sahyoun AM, Chen C, Kassan M. Ait-Aissa K, et al. Cardiovasc Diabetol. 2020 Sep 9;19(1):136. doi: 10.1186/s12933-020-01107-3. Cardiovasc Diabetol. 2020. PMID: 32907629 Free PMC article. Review. - The unfolded protein response: controlling cell fate decisions under ER stress and beyond.
Hetz C. Hetz C. Nat Rev Mol Cell Biol. 2012 Jan 18;13(2):89-102. doi: 10.1038/nrm3270. Nat Rev Mol Cell Biol. 2012. PMID: 22251901 Review. - Anticipatory UPR Activation: A Protective Pathway and Target in Cancer.
Shapiro DJ, Livezey M, Yu L, Zheng X, Andruska N. Shapiro DJ, et al. Trends Endocrinol Metab. 2016 Oct;27(10):731-741. doi: 10.1016/j.tem.2016.06.002. Epub 2016 Jun 25. Trends Endocrinol Metab. 2016. PMID: 27354311 Free PMC article. Review. - Life as a Vector of Dengue Virus: The Antioxidant Strategy of Mosquito Cells to Survive Viral Infection.
Cheng CC, Sofiyatun E, Chen WJ, Wang LC. Cheng CC, et al. Antioxidants (Basel). 2021 Mar 5;10(3):395. doi: 10.3390/antiox10030395. Antioxidants (Basel). 2021. PMID: 33807863 Free PMC article. Review. - The unfolded protein response element IRE1α senses bacterial proteins invading the ER to activate RIG-I and innate immune signaling.
Cho JA, Lee AH, Platzer B, Cross BCS, Gardner BM, De Luca H, Luong P, Harding HP, Glimcher LH, Walter P, Fiebiger E, Ron D, Kagan JC, Lencer WI. Cho JA, et al. Cell Host Microbe. 2013 May 15;13(5):558-569. doi: 10.1016/j.chom.2013.03.011. Cell Host Microbe. 2013. PMID: 23684307 Free PMC article. Retracted.
References
- Ron D, Walter P. Nat Rev Mol Cell Biol. 2007;8:519. - PubMed
- Cox JS, Walter P. Cell. 1996;87:391. - PubMed
- Sidrauski C, Walter P. Cell. 1997;90:1031. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials