Type III IFN receptor expression and functional characterisation in the pteropid bat, Pteropus alecto - PubMed (original) (raw)
Type III IFN receptor expression and functional characterisation in the pteropid bat, Pteropus alecto
Peng Zhou et al. PLoS One. 2011.
Abstract
Bats are rich reservoir hosts for a variety of viruses, many of which are capable of spillover to other susceptible mammals with lethal consequences. The ability of bats to remain asymptomatic to viral infection may be due to the rapid control of viral replication very early in the immune response through innate antiviral mechanisms. Type I and III interferons (IFNs) represent the first line of defence against viral infection in mammals, with both families of IFNs present in pteropid bats. To obtain further insight into the type III IFN system in bats, we describe the characterization of the type III IFN receptor (IFNλR) in the black flying fox, P. alecto with the characterization of IFNλR1 and IL10R2 genes that make up the type III IFN receptor complex. The bat IFNλR complex has a wide tissue distribution and at the cellular level, both epithelial and immune cells are responsive to IFN-λ treatment. Furthermore, we demonstrate that the bat IFNλR1 chain acts as a functional receptor. To our knowledge, this report represents the first description of an IFN receptor in any species of bat. The responsiveness of bat cells to IFN-λ support a role for the type III IFN system by epithelial and immune cells in bats.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Gene organization of P. alecto IFNλR1 and IL10R2 compared with that of corresponding human genes.
The intron-exon structures were predicted by nucleotide alignment of IFNλR1 or IL10R1 mRNA with the corresponding sequences in either the P. vampyrus or human genome sequences. Putative UTRs and exons are drawn as rectangles; introns are shown as dotted lines.
Figure 2. Alignment of the deduced amino acid sequence of P. alecto IFNλR1 (A) and IL10R2 (B) genes with human, mouse and horse IFNλR sequences.
Signal peptides are underlined and transmembrane regions are shaded. The conserved cysteine and tyrosine residues are in bold. Two of the tyrosine residues in IFNλR1, which are involved in meditation of STAT2 tyrosine phosphorylation in human IFNλR1 are conserved in bat IFN-λR1 and are further boxed. Dashes indicated similarity and dots indicate gaps.
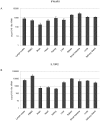
Figure 3. qRT-PCR detection of IFNλR1 (A) and IL10R2 (B) mRNA expression in P. alecto.
PBMC: peripheral blood mononuclear cells. The expression level was normalized to housekeeping gene 18s rRNA. n = 3 individual apparently healthy wild-caught bats. The error bars represent standard deviation in (A) and (B).
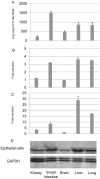
Figure 4. The responsiveness of P. alecto non-haematopoietic cell lines to IFN-λ2 treatment correlates with IFNλR1 expression and proportions of epithelial cells.
(A) Relative expression level of IFNλR1 by qRT-PCR (mean of two experiments). The expression level was normalized to the housekeeping gene 18s rRNA. (B/C) Responsiveness of P. alecto kidney, small intestine, brain, liver and lung primary cells. The histogram shows the (B) RIG-I or (C) ISG56 fold induction in response to IFN-λ2 treatment (mean of two experiments). The error bars represent standard errors in (A), (B) and (C).(D) The relative proportion of epithelial cells in the primary cell cultures as demonstrated by Western blotting using an anti-cytokeratin antibody. The housekeeping gene, GAPDH was used as a loading control.
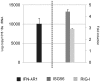
Figure 5. Responsiveness of P. alecto splenocytes to IFN-λ2 treatment and the relative expression level of IFNλR1.
Cells were treated for 6 hours before they were collected for RNA extraction and qRT-PCR analysis. The left axis shows the relative expression level of IFNλR1 (mean of three experiments). The right axis shows the ISG56 and RIG-I fold induction in response to IFN-λ2 treatment (mean of three experiments). The expression level was normalized to the housekeeping gene 18s rRNA. The error bars represent standard errors.
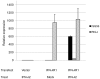
Figure 6. IFNλR1 is a functional receptor of IFN-λ2.
Bat cloned and immortalized cell line PaKiT01 cells were transfected with IFNλR1 expression plasmid or empty vector for 24 hours, followed by IFN-λ2 treatment (or mock treatment) for another 6 hours. After which, cells were collected and total RNA was extracted for qRT-PCR analysis. IFNλR1, ISG56 and RIG-I mRNA expression were tested and indicated by relative expression (mean of two experiments). The error bars represent standard errors.
Similar articles
- IRF7 in the Australian black flying fox, Pteropus alecto: evidence for a unique expression pattern and functional conservation.
Zhou P, Cowled C, Mansell A, Monaghan P, Green D, Wu L, Shi Z, Wang LF, Baker ML. Zhou P, et al. PLoS One. 2014 Aug 6;9(8):e103875. doi: 10.1371/journal.pone.0103875. eCollection 2014. PLoS One. 2014. PMID: 25100081 Free PMC article. - Bat Mx1 and Oas1, but not Pkr are highly induced by bat interferon and viral infection.
Zhou P, Cowled C, Wang LF, Baker ML. Zhou P, et al. Dev Comp Immunol. 2013 Jul-Aug;40(3-4):240-7. doi: 10.1016/j.dci.2013.03.006. Epub 2013 Mar 26. Dev Comp Immunol. 2013. PMID: 23541614 - Anti-lyssaviral activity of interferons κ and ω from the serotine bat, Eptesicus serotinus.
He X, Korytař T, Schatz J, Freuling CM, Müller T, Köllner B. He X, et al. J Virol. 2014 May;88(10):5444-54. doi: 10.1128/JVI.03403-13. Epub 2014 Feb 26. J Virol. 2014. PMID: 24574413 Free PMC article. - Fundamental Characteristics of Bat Interferon Systems.
Clayton E, Munir M. Clayton E, et al. Front Cell Infect Microbiol. 2020 Dec 11;10:527921. doi: 10.3389/fcimb.2020.527921. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33363045 Free PMC article. Review. - Genomic and biological variation in bat IFNs: An antiviral treatment approach.
Al-Eitan L, Mihyar A, Zhang L, Bisht P, Jaenisch R. Al-Eitan L, et al. Rev Med Virol. 2024 Jan;34(1):e2488. doi: 10.1002/rmv.2488. Epub 2023 Nov 3. Rev Med Virol. 2024. PMID: 37921610 Review.
Cited by
- IRF7 in the Australian black flying fox, Pteropus alecto: evidence for a unique expression pattern and functional conservation.
Zhou P, Cowled C, Mansell A, Monaghan P, Green D, Wu L, Shi Z, Wang LF, Baker ML. Zhou P, et al. PLoS One. 2014 Aug 6;9(8):e103875. doi: 10.1371/journal.pone.0103875. eCollection 2014. PLoS One. 2014. PMID: 25100081 Free PMC article. - Henipavirus Immune Evasion and Pathogenesis Mechanisms: Lessons Learnt from Natural Infection and Animal Models.
Lawrence P, Escudero-Pérez B. Lawrence P, et al. Viruses. 2022 Apr 29;14(5):936. doi: 10.3390/v14050936. Viruses. 2022. PMID: 35632678 Free PMC article. Review. - Molecular, ecological, and behavioral drivers of the bat-virus relationship.
Gonzalez V, Banerjee A. Gonzalez V, et al. iScience. 2022 Aug 19;25(8):104779. doi: 10.1016/j.isci.2022.104779. Epub 2022 Jul 20. iScience. 2022. PMID: 35875684 Free PMC article. Review. - Identification, Characterization, and Developmental Expression Pattern of Type III Interferon Receptor Gene in the Chinese Goose.
Zhou Q, Chen S, Qi Y, Zhou H, Wang M, Jia R, Zhu D, Liu M, Liu F, Chen X, Zhou X, Cheng A. Zhou Q, et al. Biomed Res Int. 2015;2015:186274. doi: 10.1155/2015/186274. Epub 2015 May 10. Biomed Res Int. 2015. PMID: 26064884 Free PMC article. - Functional characterization of canine interferon-lambda.
Fan W, Xu L, Ren L, Qu H, Li J, Liang J, Liu W, Yang L, Luo T. Fan W, et al. J Interferon Cytokine Res. 2014 Nov;34(11):848-57. doi: 10.1089/jir.2014.0009. Epub 2014 Jun 20. J Interferon Cytokine Res. 2014. PMID: 24950142 Free PMC article.
References
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. - PubMed
- Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–8. - PubMed
- Langer JA, Cutrone EC, Kotenko S. The Class II cytokine receptor (CRF2) family: overview and patterns of receptor-ligand interactions. Cytokine and Growth Factor Reviews. 2004;15:33–48. - PubMed
- Ank N, Paludan SR. Type III IFNs: new layers of complexity in innate antiviral immunity. Biofactors. 2009;35:82–7. - PubMed
- Witte K, Witte E, Sabat R, Wolk K. IL-28A, IL-28B, and IL-29: Promising cytokines with type I interferon-like properties. Cytokine Growth Factor Rev. 2010;21:237–251. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources