Mitochondrial genome sequencing helps show the evolutionary mechanism of mitochondrial genome formation in Brassica - PubMed (original) (raw)
Mitochondrial genome sequencing helps show the evolutionary mechanism of mitochondrial genome formation in Brassica
Shengxin Chang et al. BMC Genomics. 2011.
Abstract
Background: Angiosperm mitochondrial genomes are more complex than those of other organisms. Analyses of the mitochondrial genome sequences of at least 11 angiosperm species have showed several common properties; these cannot easily explain, however, how the diverse mitotypes evolved within each genus or species. We analyzed the evolutionary relationships of Brassica mitotypes by sequencing.
Results: We sequenced the mitotypes of cam (Brassica rapa), ole (B. oleracea), jun (B. juncea), and car (B. carinata) and analyzed them together with two previously sequenced mitotypes of B. napus (pol and nap). The sizes of whole single circular genomes of cam, jun, ole, and car are 219,747 bp, 219,766 bp, 360,271 bp, and 232,241 bp, respectively. The mitochondrial genome of ole is largest as a resulting of the duplication of a 141.8 kb segment. The jun mitotype is the result of an inherited cam mitotype, and pol is also derived from the cam mitotype with evolutionary modifications. Genes with known functions are conserved in all mitotypes, but clear variation in open reading frames (ORFs) with unknown functions among the six mitotypes was observed. Sequence relationship analysis showed that there has been genome compaction and inheritance in the course of Brassica mitotype evolution.
Conclusions: We have sequenced four Brassica mitotypes, compared six Brassica mitotypes and suggested a mechanism for mitochondrial genome formation in Brassica, including evolutionary events such as inheritance, duplication, rearrangement, genome compaction, and mutation.
Figures
Figure 1
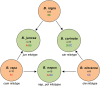
Cytogenetic relationships of six cultivated Brassica species as depicted by U's triangle [20]. U's triangle illustrates the evolutionary relationship between three cultivated elementary species (B. rapa, B. oleracea, and B. nigra) and three amphiploid species (B. napus, B. juncea, and B. carinata). Chromosome numbers, nuclear genome types and mitotypes are shown inside or outside the circle for each species.
Figure 2
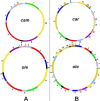
Large repeats exist in the six mitotypes. RB, R1, R, and R2 denote repeats of more than 2 kb. RB and R1 are shared by the six mitotypes, but their copy numbers vary.
Figure 3
Rearrangements of Brassica mitochondrial genomes. Syntenic regions > 2 kb are shown. (A) Rearrangement of the cam mitochondrial genome with the ole mitotype as a reference. (B) Rearrangement of the car mitochondrial genome with the ole mitotype as a reference. The numbers refer to the syntenic regions derived from a paired comparison. Highly or completely homologous regions are indicated by color.
Figure 4
Short repeats associated with changes in the mitochondrial genome of Brassica. The orientation of the sequence is shown by an arrow. (A) Repeat Q is possibly related to the influence of the rearrangement of syntenic regions 7, 2, 5, 6, 8, and 1 of the ole and cam mitotypes. (B) Repeat P may be related to the rearrangement of regions 16 and 1 in the ole and car mitotypes.
Figure 5
Clustering tree of Brassica mitotypes. It's according to the distance based on indels > 400 bp and SNPs (see Additional file 1, Tables S4 and S5).
Figure 6
Hypothesis regarding the evolution of six cultivated Brassica mitotypes. Diverse Brassica mitotypes are hypothesized to have evolved from an expanded ancestral parent mitotype and formed through mitochondrial genome speciation and compaction. Jun (B. juncea) is derived from the cam mitotype and pol and nap from a primary mitotype very similar to the cam mitotype, without deletion of the CMS-related orf224 gene region (4.4 kb) or its homolog orf222. The maternal mitotype from which car is derived is unclear (dotted lines). Three mitotypes for the elementary species are also hypothesized to be compaction forms from the large ancestral mitotype.
Similar articles
- Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus L.
Chen J, Guan R, Chang S, Du T, Zhang H, Xing H. Chen J, et al. PLoS One. 2011 Mar 10;6(3):e17662. doi: 10.1371/journal.pone.0017662. PLoS One. 2011. PMID: 21423700 Free PMC article. - Complete sequence of heterogenous-composition mitochondrial genome (Brassica napus) and its exogenous source.
Wang J, Jiang J, Li X, Li A, Zhang Y, Guan R, Wang Y. Wang J, et al. BMC Genomics. 2012 Nov 28;13:675. doi: 10.1186/1471-2164-13-675. BMC Genomics. 2012. PMID: 23190559 Free PMC article. - Identification of different cytoplasms based on newly developed mitotype-specific markers for marker-assisted selection breeding in Brassica napus L.
Heng S, Chen F, Wei C, Hu K, Yang Z, Wen J, Yi B, Ma C, Tu J, Si P, Fu T, Shen J. Heng S, et al. Plant Cell Rep. 2017 Jun;36(6):901-909. doi: 10.1007/s00299-017-2121-4. Epub 2017 Mar 6. Plant Cell Rep. 2017. PMID: 28265748 - Comparative mitochondrial genome analysis reveals the evolutionary rearrangement mechanism in Brassica.
Yang J, Liu G, Zhao N, Chen S, Liu D, Ma W, Hu Z, Zhang M. Yang J, et al. Plant Biol (Stuttg). 2016 May;18(3):527-36. doi: 10.1111/plb.12414. Epub 2015 Nov 29. Plant Biol (Stuttg). 2016. PMID: 27079962
Cited by
- Assembly and comparative analysis of the first complete mitochondrial genome of zicaitai (Brassica rapa var. Purpuraria): insights into its genetic architecture and evolutionary relationships.
Xiao W, Wu X, Zhou X, Zhang J, Huang J, Dai X, Ren H, Xu D. Xiao W, et al. Front Plant Sci. 2024 Oct 10;15:1475064. doi: 10.3389/fpls.2024.1475064. eCollection 2024. Front Plant Sci. 2024. PMID: 39450086 Free PMC article. - Assembly and Comparative Analysis of the Complete Mitochondrial Genome of Saussurea inversa (Asteraceae).
Dai W, Ju X, Shi G, He T. Dai W, et al. Genes (Basel). 2024 Aug 14;15(8):1074. doi: 10.3390/genes15081074. Genes (Basel). 2024. PMID: 39202433 Free PMC article. - A Systematic Review and Developmental Perspective on Origin of CMS Genes in Crops.
Zhang X, Ding Z, Lou H, Han R, Ma C, Yang S. Zhang X, et al. Int J Mol Sci. 2024 Jul 31;25(15):8372. doi: 10.3390/ijms25158372. Int J Mol Sci. 2024. PMID: 39125940 Free PMC article. Review. - De Novo Hybrid Assembly Unveils Multi-Chromosomal Mitochondrial Genomes in Ludwigia Species, Highlighting Genomic Recombination, Gene Transfer, and RNA Editing Events.
Doré G, Barloy D, Barloy-Hubler F. Doré G, et al. Int J Mol Sci. 2024 Jul 2;25(13):7283. doi: 10.3390/ijms25137283. Int J Mol Sci. 2024. PMID: 39000388 Free PMC article. - Assembly and comparative analysis of the complete mitochondrial genome of Brassica rapa var. Purpuraria.
Gong Y, Xie X, Zhou G, Chen M, Chen Z, Li P, Huang H. Gong Y, et al. BMC Genomics. 2024 Jun 1;25(1):546. doi: 10.1186/s12864-024-10457-1. BMC Genomics. 2024. PMID: 38824587 Free PMC article.
References
- Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, Hirai A, Kadowaki K. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics. 2002;268(4):434–445. doi: 10.1007/s00438-002-0767-1. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous