Normalization, testing, and false discovery rate estimation for RNA-sequencing data - PubMed (original) (raw)
Normalization, testing, and false discovery rate estimation for RNA-sequencing data
Jun Li et al. Biostatistics. 2012 Jul.
Abstract
We discuss the identification of genes that are associated with an outcome in RNA sequencing and other sequence-based comparative genomic experiments. RNA-sequencing data take the form of counts, so models based on the Gaussian distribution are unsuitable. Moreover, normalization is challenging because different sequencing experiments may generate quite different total numbers of reads. To overcome these difficulties, we use a log-linear model with a new approach to normalization. We derive a novel procedure to estimate the false discovery rate (FDR). Our method can be applied to data with quantitative, two-class, or multiple-class outcomes, and the computation is fast even for large data sets. We study the accuracy of our approaches for significance calculation and FDR estimation, and we demonstrate that our method has potential advantages over existing methods that are based on a Poisson or negative binomial model. In summary, this work provides a pipeline for the significance analysis of sequencing data.
Figures
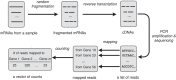
Fig 1.
Pipeline for a typical RNA-Seq experiment. Firstly, mRNA is randomly fragmented into small pieces. These small pieces are then reverse transcribed into a cDNA library by random priming. Then this cDNA library is amplified via PCR and sequenced by a sequencing machine, producing a list of reads. These reads are mapped to a known transcriptome which consists of m genes, and the number of reads mapping to each gene is used as a measure of gene expression. Thus, the results of an RNA-Seq experiment are summarized by a vector of m counts.
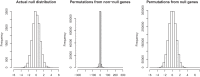
Fig 2.
Histograms of score statistics for simulated data with a two-class outcome. Here, we use the signed version of the score statistic for a more clear display. The permutation distribution of the non-null genes (middle) is much wider than the permutation distribution of the null genes (right), which is very similar to the true null distribution (left).
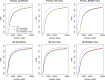
Fig 3.
FDR curves for simulated (top three panels) Poisson distributed data and (bottom three panels) negative binomial distributed data. The left, middle, and right panels show results from data with different types of outcome (averaged over 100 simulations). The solid curves show the true FDRs; the broken curves are estimates. All curves are based on the score statistic (3.8 and 3.9) but use different methods to estimate FDRs: PoissonSeq, the usual permutation plug-in method (permutation), and the theoretical p-value method (theoretical p-value). The true FDR curve, which is the same for all three procedures, is also shown. In the Poisson case, both PoissonSeq and the theoretical p-value method give much more accurate FDR estimates than the usual permutation plug-in method. In the negative binomial case, the PoissonSeq estimate of FDR is much more accurate than the other two estimates.
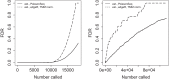
Fig 4.
FDR curves for simulated (left) Poisson-distributed data and (right) negative binomial distributed data. The solid curves show the true FDRs; the broken curves are estimates. These are results (averaged over 100 simulations) on data with two-class outcome using different methods: our method (PoissonSeq), SAM applied to the square root of total-count normalized data (SAM), the method proposed by Marioni and others (2008) (LRT on Poisson), edgeR with the default total-count normalization (edgeR, total-count norm.), and edgeR with TMM normalization (edgeR, TMM norm). In the Poisson case, we see that only edgeR with TMM normalization and our PoissonSeq method yield accurate FDR estimates. PoissonSeq and edgeR with TMM normalization also yield much lower true FDRs than the other methods. In the negative binomial case, only the results using PoissonSeq and edgeR with TMM normalization are shown. We see that the true FDR curves of the two methods are almost the same, while our estimate is more accurate.
Fig 5.
FDR curves estimated by PoissonSeq and edgeR with TMM normalization for two data sets: (left) the data set from Marioni and others (2008) and (right) the data set from 't Hoen and others (2008).
Similar articles
- Finding consistent patterns: a nonparametric approach for identifying differential expression in RNA-Seq data.
Li J, Tibshirani R. Li J, et al. Stat Methods Med Res. 2013 Oct;22(5):519-36. doi: 10.1177/0962280211428386. Epub 2011 Nov 28. Stat Methods Med Res. 2013. PMID: 22127579 Free PMC article. - Accuracy of RNA-Seq and its dependence on sequencing depth.
Cai G, Li H, Lu Y, Huang X, Lee J, Müller P, Ji Y, Liang S. Cai G, et al. BMC Bioinformatics. 2012;13 Suppl 13(Suppl 13):S5. doi: 10.1186/1471-2105-13-S13-S5. Epub 2012 Aug 24. BMC Bioinformatics. 2012. PMID: 23320920 Free PMC article. - Is this the right normalization? A diagnostic tool for ChIP-seq normalization.
Angelini C, Heller R, Volkinshtein R, Yekutieli D. Angelini C, et al. BMC Bioinformatics. 2015 May 9;16:150. doi: 10.1186/s12859-015-0579-z. BMC Bioinformatics. 2015. PMID: 25957089 Free PMC article. - Sample size calculation while controlling false discovery rate for differential expression analysis with RNA-sequencing experiments.
Bi R, Liu P. Bi R, et al. BMC Bioinformatics. 2016 Mar 31;17:146. doi: 10.1186/s12859-016-0994-9. BMC Bioinformatics. 2016. PMID: 27029470 Free PMC article. - cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate.
Klambauer G, Schwarzbauer K, Mayr A, Clevert DA, Mitterecker A, Bodenhofer U, Hochreiter S. Klambauer G, et al. Nucleic Acids Res. 2012 May;40(9):e69. doi: 10.1093/nar/gks003. Epub 2012 Feb 1. Nucleic Acids Res. 2012. PMID: 22302147 Free PMC article.
Cited by
- RNA-Seq Count Data Modelling by Grey Relational Analysis and Nonparametric Gaussian Process.
Nguyen T, Bhatti A, Yang S, Nahavandi S. Nguyen T, et al. PLoS One. 2016 Oct 26;11(10):e0164766. doi: 10.1371/journal.pone.0164766. eCollection 2016. PLoS One. 2016. PMID: 27783633 Free PMC article. - Epitranscriptome analysis of NAD-capped RNA by spike-in-based normalization and prediction of chronological age.
Li D, Ge S, Liu Y, Pan M, Wang X, Han G, Zou S, Liu R, Niu K, Zhao C, Liu N, Qu L. Li D, et al. iScience. 2023 Nov 22;26(12):108558. doi: 10.1016/j.isci.2023.108558. eCollection 2023 Dec 15. iScience. 2023. PMID: 38094247 Free PMC article. - Single-cell RNA sequencing technologies and bioinformatics pipelines.
Hwang B, Lee JH, Bang D. Hwang B, et al. Exp Mol Med. 2018 Aug 7;50(8):1-14. doi: 10.1038/s12276-018-0071-8. Exp Mol Med. 2018. PMID: 30089861 Free PMC article. Review. - Differential microRNA expression profiles determined by next-generation sequencing in three fulvestrant-resistant human breast cancer cell lines.
Guo J, He K, Zeng H, Shi Y, Ye P, Zhou Q, Pan Z, Long X. Guo J, et al. Oncol Lett. 2019 Apr;17(4):3765-3776. doi: 10.3892/ol.2019.10061. Epub 2019 Feb 21. Oncol Lett. 2019. PMID: 30930984 Free PMC article. - cdev: a ground-truth based measure to evaluate RNA-seq normalization performance.
Tran DT, Might M. Tran DT, et al. PeerJ. 2021 Oct 4;9:e12233. doi: 10.7717/peerj.12233. eCollection 2021. PeerJ. 2021. PMID: 34707933 Free PMC article.
References
- Agresti A. Categorical Data Analysis. 2nd edition. New York: Wiley; 2002.
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;85:289–300.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources