Genomic analysis identifies association of Fusobacterium with colorectal carcinoma - PubMed (original) (raw)
doi: 10.1101/gr.126573.111. Epub 2011 Oct 18.
Dirk Gevers, Chandra Sekhar Pedamallu, Monia Michaud, Fujiko Duke, Ashlee M Earl, Akinyemi I Ojesina, Joonil Jung, Adam J Bass, Josep Tabernero, José Baselga, Chen Liu, Ramesh A Shivdasani, Shuji Ogino, Bruce W Birren, Curtis Huttenhower, Wendy S Garrett, Matthew Meyerson
Affiliations
- PMID: 22009990
- PMCID: PMC3266036
- DOI: 10.1101/gr.126573.111
Genomic analysis identifies association of Fusobacterium with colorectal carcinoma
Aleksandar D Kostic et al. Genome Res. 2012 Feb.
Abstract
The tumor microenvironment of colorectal carcinoma is a complex community of genomically altered cancer cells, nonneoplastic cells, and a diverse collection of microorganisms. Each of these components may contribute to carcinogenesis; however, the role of the microbiota is the least well understood. We have characterized the composition of the microbiota in colorectal carcinoma using whole genome sequences from nine tumor/normal pairs. Fusobacterium sequences were enriched in carcinomas, confirmed by quantitative PCR and 16S rDNA sequence analysis of 95 carcinoma/normal DNA pairs, while the Bacteroidetes and Firmicutes phyla were depleted in tumors. Fusobacteria were also visualized within colorectal tumors using FISH. These findings reveal alterations in the colorectal cancer microbiota; however, the precise role of Fusobacteria in colorectal carcinoma pathogenesis requires further investigation.
Figures
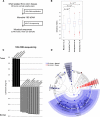
Figure 1.
Whole-genome sequencing analysis of the colorectal cancer microbiome. (A) Schematic of experimental and computational whole-genome sequencing analysis workflow. (B) Hierarchical clustering of phylotype relative abundance measurements demonstrates that microbial composition of tumor/normal pairs within individuals is more highly correlated than tumor/tumor pairs or normal/normal pairs from different individuals. (Green) Normal samples; (purple) tumors. (C) Linear discriminant analysis (LDA) coupled with effect size measurements identifies Fusobacterium as the most differentially abundant taxon in colon tumor versus normal specimens by whole-genome sequencing in nine individuals. Tumor-enriched taxa are indicated with a positive LDA score (black), and taxa enriched in normal tissue have a negative score (gray). Only taxa meeting an LDA significant threshold of 1.8 are shown. (D) Percent relative abundance for the genus Fusobacterium is depicted across all samples in the order of the labels in B, demonstrating a tumor-enrichment in most individuals.
Figure 2.
16S rDNA sequencing analysis of the colorectal cancer microbiome. (A) Schematic of experimental and computational 16S rDNA sequencing analysis workflow. (B) Beta-diversity distances calculated using phylotype relative abundance measurements between all pairs of samples demonstrate that the microbial composition of tumor/normal pairs within individuals is more highly correlated than tumor/tumor pairs, normal/normal pairs, or tumor/normal pairs from different individuals. (C) Linear discriminant analysis (LDA) coupled with effect size measurements identifies Fusobacterium as the most differentially abundant taxon in colon tumor versus normal specimens by 16S rDNA sequencing in 95 individuals. Tumor-enriched taxa are indicated with a positive LDA score (black), and taxa enriched in normal tissue have a negative score (gray). Only taxa meeting an LDA significant threshold of 4.2 are shown. (D) A cladogram representation of data in C. (Red) Tumor-enriched taxa; (blue) taxa enriched in normal tissue. The brightness of each dot is proportional to its effect size.
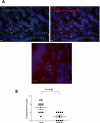
Figure 3.
Fluorescence in situ hybridization (FISH) detects enrichment of Fusobacteria in colorectal tumors. (A) FISH using an Oregon-Green 488-conjugated ‘‘universal bacterial'' 16S rDNA-directed oligonucleotide probe (EUB338, green) (top left); and Cy3-conjugated Fusobacterium (FUSO, red) (top right and bottom center) 16S rDNA-direct oligonucleotide probe demonstrates the presence of bacteria and Fusobacterium within the colonic mucosa of colorectal tumor samples. Representative images are shown with a 10-μm scale bar in the lower corner of each panel; white arrowheads mark bacteria. Epithelial cell nuclei were stained with DAPI. (B) To determine whether Fusobacterium was enriched in tumor versus normal pairs, three random 40× fields were chosen for scoring by an observer blind to tumor/normal status, using selection criteria of mucosal tissue depth and a minimum of five bacteria visualized by the EUB338 probe per field. Each dot represents data from either a tumor or normal sample from nine tumor/normal paired cases. The mean, SEM, and _P_-values (calculated by a Wilcoxon matched-pairs signed rank test) are shown.
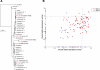
Figure 4.
Phylogenetic analysis identifies several Fusobacterium species in human colon cancer tissues. (A) Approximately-maximum-likelihood phylogenetic trees were constructed on the V3–V5 region of the 16S rDNA gene using 31 reference Fusobacterium species along with the five most prominent OTUs identified in colon cancer specimens (indicated in red). Nodes that have bootstrap support above 50% and 75% are indicated with a white and black dot, respectively. The mean percent relative abundance in tumor (T) and normal (N) of each OTU is indicated in parentheses. The full names of the reference strains appear in Supplemental Table S6. (B) The abundance of the indicated OTU relative to all other phylotypes in a given specimen is shown for the two most abundant Fusobacterium OTUs in tumors (_x_-axis) and normal colon tissue (_y_-axis); each point represents tumor and normal abundance data for a different individual. The lower-right quadrant of the graph highlights the substantial proportion of patients for whom the Fusobacterium abundance is >10% in tumors but <10% in the matched normal.
Comment in
- Colorectal cancer: Fusobacterium nucleatum found in colon cancer tissue--could an infection cause colorectal cancer?
Ray K. Ray K. Nat Rev Gastroenterol Hepatol. 2011 Nov 15;8(12):662. doi: 10.1038/nrgastro.2011.208. Nat Rev Gastroenterol Hepatol. 2011. PMID: 22083120 No abstract available.
Similar articles
- Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma.
Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Castellarin M, et al. Genome Res. 2012 Feb;22(2):299-306. doi: 10.1101/gr.126516.111. Epub 2011 Oct 18. Genome Res. 2012. PMID: 22009989 Free PMC article. - Role of Fusobacteria in the serrated pathway of colorectal carcinogenesis.
Park CH, Han DS, Oh YH, Lee AR, Lee YR, Eun CS. Park CH, et al. Sci Rep. 2016 Apr 29;6:25271. doi: 10.1038/srep25271. Sci Rep. 2016. PMID: 27125587 Free PMC article. - Mucosa-associated microbiota signature in colorectal cancer.
Gao R, Kong C, Huang L, Li H, Qu X, Liu Z, Lan P, Wang J, Qin H. Gao R, et al. Eur J Clin Microbiol Infect Dis. 2017 Nov;36(11):2073-2083. doi: 10.1007/s10096-017-3026-4. Epub 2017 Jun 9. Eur J Clin Microbiol Infect Dis. 2017. PMID: 28600626 - Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer.
Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H, Maruyama R, Imai K, Yamamoto H, Shinomura Y. Nosho K, et al. World J Gastroenterol. 2016 Jan 14;22(2):557-66. doi: 10.3748/wjg.v22.i2.557. World J Gastroenterol. 2016. PMID: 26811607 Free PMC article. Review. - Fusobacterium nucleatum: an emerging bug in colorectal tumorigenesis.
Bashir A, Miskeen AY, Bhat A, Fazili KM, Ganai BA. Bashir A, et al. Eur J Cancer Prev. 2015 Sep;24(5):373-85. doi: 10.1097/CEJ.0000000000000116. Eur J Cancer Prev. 2015. PMID: 25569450 Review.
Cited by
- Gut bacteria: an etiological agent in human pathological conditions.
Islam MM, Mahbub NU, Hong ST, Chung HJ. Islam MM, et al. Front Cell Infect Microbiol. 2024 Oct 8;14:1291148. doi: 10.3389/fcimb.2024.1291148. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39439902 Free PMC article. Review. - The gastrointestinal microbiota and colorectal cancer.
Keku TO, Dulal S, Deveaux A, Jovov B, Han X. Keku TO, et al. Am J Physiol Gastrointest Liver Physiol. 2015 Mar 1;308(5):G351-63. doi: 10.1152/ajpgi.00360.2012. Epub 2014 Dec 24. Am J Physiol Gastrointest Liver Physiol. 2015. PMID: 25540232 Free PMC article. Review. - From the Colon to the Liver: How Gut Microbiota May Influence Colorectal Cancer Metastatic Potential.
Mignini I, Piccirilli G, Galasso L, Termite F, Esposto G, Ainora ME, Gasbarrini A, Zocco MA. Mignini I, et al. J Clin Med. 2024 Jan 12;13(2):420. doi: 10.3390/jcm13020420. J Clin Med. 2024. PMID: 38256554 Free PMC article. Review. - The microbiota and microbiome in aging: potential implications in health and age-related diseases.
Zapata HJ, Quagliarello VJ. Zapata HJ, et al. J Am Geriatr Soc. 2015 Apr;63(4):776-81. doi: 10.1111/jgs.13310. Epub 2015 Apr 8. J Am Geriatr Soc. 2015. PMID: 25851728 Free PMC article. - Association of plasma endotoxin, inflammatory cytokines and risk of colorectal adenomas.
Kang M, Edmundson P, Araujo-Perez F, McCoy AN, Galanko J, Keku TO. Kang M, et al. BMC Cancer. 2013 Feb 26;13:91. doi: 10.1186/1471-2407-13-91. BMC Cancer. 2013. PMID: 23442743 Free PMC article.
References
- Bachrach G, Ianculovici C, Naor R, Weiss EI 2005. Fluorescence-based measurements of Fusobacterium nucleatum coaggregation and of fusobacterial attachment to mammalian cells. FEMS Microbiol Lett 248: 235–240 - PubMed
- Boutaga K, van Winkelhoff AJ, Vandenbroucke-Grauls CM, Savelkoul PH 2005. Periodontal pathogens: A quantitative comparison of anaerobic culture and real-time PCR. FEMS Immunol Med Microbiol 45: 191–199 - PubMed
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266: 1865–1869 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01CA154426/CA/NCI NIH HHS/United States
- UL1 TR000064/TR/NCATS NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- K08 CA134931/CA/NCI NIH HHS/United States
- U54HG003067/HG/NHGRI NIH HHS/United States
- P50CA127003/CA/NCI NIH HHS/United States
- R01 CA154426/CA/NCI NIH HHS/United States
- RC2CA148317/CA/NCI NIH HHS/United States
- P50 CA127003/CA/NCI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- RC2 CA148317/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical