Platelet-derived growth factor-C (PDGF-C) activation by serine proteases: implications for breast cancer progression - PubMed (original) (raw)
Platelet-derived growth factor-C (PDGF-C) activation by serine proteases: implications for breast cancer progression
Newton J Hurst Jr et al. Biochem J. 2012.
Abstract
The PDGF (platelet-derived growth factor) family members are potent mitogens for cells of mesenchymal origin and serve as important regulators of cell migration, survival, apoptosis and transformation. Tumour-derived PDGF ligands are thought to function in both autocrine and paracrine manners, activating receptors on tumour and surrounding stromal cells. PDGF-C and -D are secreted as latent dimers, unlike PDGF-A and -B. Cleavage of the CUB domain from the PDGF-C and -D dimers is required for their biological activity. At present, little is known about the proteolytic processing of PDGF-C, the rate-limiting step in the regulation of PDGF-C activity. In the present study we show that the breast carcinoma cell line MCF7, engineered to overexpress PDGF-C, produces proteases capable of cleaving PDGF-C to its active form. Increased PDGF-C expression enhances cell proliferation, anchorage-independent cell growth and tumour cell motility by autocrine signalling. In addition, MCF7-produced PDGF-C induces fibroblast cell migration in a paracrine manner. Interestingly, PDGF-C enhances tumour cell invasion in the presence of fibroblasts, suggesting a role for tumour-derived PDGF-C in tumour-stromal interactions. In the present study, we identify tPA (tissue plasminogen activator) and matriptase as major proteases for processing of PDGF-C in MCF7 cells. In in vitro studies, we also show that uPA (urokinase-type plasminogen activator) is able to process PDGF-C. Furthermore, by site-directed mutagenesis, we identify the cleavage site for these proteases in PDGF-C. Lastly, we provide evidence suggesting a two-step proteolytic processing of PDGF-C involving creation of a hemidimer, followed by GFD-D (growth factor domain dimer) generation.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Figures
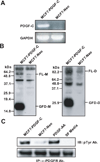
Figure 1. Expression and processing of platelet-derived growth factor-C (PDGF-C) in MCF7
A. PDGF-C levels in MCF7-PDGF-C (Lane 1) and -Neo (Lane 2) were examined by RT-PCR. B. Serum-free (SF) conditioned media (CM) collected from MCF7-PDGF C or -Neo cells were analyzed by immunoblot analysis using anti-PDGF-C growth factor domain Ab in reducing (left panel) and non-reducing (right panel) conditions. FL-M, full-length monomer; GFD-M, growth factor domain monomer; FL-D, full-length dimer; GFD-D, growth factor domain dimer. C. Serum-starved NIH3T3 fibroblasts were stimulated for 10 min with CM collected from MCF7-PDGF-C (Lane 1) or MCF7-Neo (Lane 2) cells, SF media with 20ng/ml PDGF-AA (Lane 3, positive control), or SF media (Lane 4, negative control). Cell lysates were immunoprecipitated using anti-PDGF-C Ab and activated α-PDGFR was detected by immunoblot using a pTyr Ab.
Figure 2. In vitro transformative properties of PDGF-C in MCF7 cells
A (paracrine signaling). A scratch migration assay of NIH3T3 was performed in the presence of conditioned media collected from MCF7-neo or –PDGF-C cells. Using NIH ImageJ, closure of the gap was quantified as a percentage of cleared area remaining at time 0, 8, and 16 from three independent experiments (top panel). Representative 40X images of time 0 and 16 hours are displayed (bottom panel). B and C (autocrine signaling). Anchorage-independent growth and proliferation of MCF7-neo or –PDGF-C cells were assessed by a soft agar colony formation assay (B) and WST-1 cell proliferation assay (C), respectively. Positive colonies were quantified from three separate experiments using Optronix GelCount (Oxford, England) in panel B and cell proliferation was quantified from three independent WST-1 assays in panel C. D (autocrine and double paracrine signaling). Matrigel invasion assay of MCF7-neo or –PDGF-C cells through a modified Boyden chamber was performed in the absence or presence of NIH3T3 fibroblasts in the bottom chamber. Quantitation is averaged results of three separate experiments. *p<0.05.
Figure 3. PDGF-C is processed by serine proteases, specifically tPA and uPA
A. MCF7-PDGF-C cells were incubated in SF media with the various class specific inhibitors for 48h; the collected CM was resolved under reducing SDS-PAGE and immunoblotted with an anti-PDGF-C GFD Ab. B. Left panel, MCF7-PDGF-C cells were incubated with SF media without or with PAI-1. Right panel, MCF7-PDGF-C (Lanes 1 and 3) or MCF7-neo (Lanes 2 and 4) cells were incubated with SF media containing tPA or uPA specific inhibitors for 48h; the collected CM was resolved under reducing SDS-PAGE and immunoblotted with an anti-PDGF-C GFD Ab. C. MCF7-PDGF-C (Lane 3) and MCF7-Neo (Lane 4) CM was run in a plasminogen-dependent zymogram with r-tPA (Lane 1) and r-uPA (Lane 2) serving as positive activity controls. D. rPDGF-C was generated by co-infecting/transfecting CV-1 cells with vaccinia virus and the pTF7-PDGF-C: HIS construct. After 48h of serum-starvation the CM was collected. This CM was concentrated using Ni-NTA agarose beads overnight. After washing of the beads, rPDGF-C was eluted with 10mM EDTA then rPDGF-C was incubated with tPA or uPA overnight. Finally, the resultant products were resolved by SDS-PAGE and immunoblotted with an anti-PDGF-C GFD Ab. E. rPDGF-C was incubated with tPA, uPA, streptokinase, and streptokinase+glu-plasminogen overnight, and the resultant products were resolved by SDS-PAGE and immunoblotted with an anti-PDGF-C GFD Ab.
Figure 4. Mutational analysis identifies tPA and uPA cleavage sites
A. Comparison of sequence alignments between human PDGF-C, murine PDGF-C, and human PDGF-D. Mutagenesis sites are marked with asterisks and the tPA/uPA substrate specificity in the P1–P4 residues are shown. B. Western blot analysis of PDGF-C wild-type (WT), PDGF-C K225A, PDGF-C R231A/234A, and PDGF-C K225A/R231A/R234A mutants under reducing (left panel) and non-reducing (right panel) conditions when incubated with 100nM of tPA 4h at 37°C in the presence of fibrinogen fragments. C. Western blot analysis of PDGF-C WT, PDGF-C K225A, PDGF-C R231A/R234A, and PDGF-C K225A/R231A/R234A under reducing (left panel) and non-reducing (right panel) conditions when incubated with 100nM of uPA overnight at 37°C.
Figure 5. Matriptase is capable of processing PDGF-C
A. Western blot analysis of MCF7-PDGF-C and -Neo CM and lysates for matriptase expression. Blot was probed with anti-matriptase Ab (kind gift of Dr. C-Y Lin, University of Maryland) under the non-reducing condition. B. rPDGF-C WT incubated with varying concentrations of matriptase 2h at 37°C and then resolved on SDS-PAGE under reducing conditions C. rPDGF-C WT and mutants were incubated with 1nM matriptase for 2h at 37°C and then resolved on SDS-PAGE under reducing conditions (top panel) or non-reducing conditions (bottom panel). D. rPDGF-C WT or R231A/R234A were incubated 2h at 37°C with 1nM matriptase before being suspended in SF DMEM/F12 media and then added to NIH3T3 cells for 10 min. Subsequently, the lysates from these cells were analyzed by western blot for the presence of phospho-β-PDGFR. Total β-PDGFR was used as a loading control. MCF7-PDGF-C cells were treated with the matriptase inhibitor, HAI-1, and PDGF-C processing (E) and biological activity (F) were monitored. In the same gel, an unnecessary lane separating SFM and NT treatment was removed in panel E. A white line is drawn to demarcate this change. G. Conditioned media from (E) were utilized to assess NIH3T3 cell migration. * and ** p<0.05.
Similar articles
- Platelet-derived growth factor D is activated by urokinase plasminogen activator in prostate carcinoma cells.
Ustach CV, Kim HR. Ustach CV, et al. Mol Cell Biol. 2005 Jul;25(14):6279-88. doi: 10.1128/MCB.25.14.6279-6288.2005. Mol Cell Biol. 2005. PMID: 15988036 Free PMC article. - The uPA/uPAR system regulates the bioavailability of PDGF-DD: implications for tumour growth.
Ehnman M, Li H, Fredriksson L, Pietras K, Eriksson U. Ehnman M, et al. Oncogene. 2009 Jan 29;28(4):534-44. doi: 10.1038/onc.2008.410. Epub 2008 Nov 10. Oncogene. 2009. PMID: 18997817 - Tissue-type plasminogen activator is not necessary for platelet-derived growth factor-c activation.
Riehle KJ, Johnson MM, Johansson F, Bauer RL, Hayes BJ, Gilbertson DG, Haran AC, Fausto N, Campbell JS. Riehle KJ, et al. Biochim Biophys Acta. 2014 Feb;1842(2):318-25. doi: 10.1016/j.bbadis.2013.11.013. Epub 2013 Nov 19. Biochim Biophys Acta. 2014. PMID: 24269585 Free PMC article. - Structural and functional specificities of PDGF-C and PDGF-D, the novel members of the platelet-derived growth factors family.
Reigstad LJ, Varhaug JE, Lillehaug JR. Reigstad LJ, et al. FEBS J. 2005 Nov;272(22):5723-41. doi: 10.1111/j.1742-4658.2005.04989.x. FEBS J. 2005. PMID: 16279938 Review. - PDGF-D signaling: a novel target in cancer therapy.
Wang Z, Kong D, Li Y, Sarkar FH. Wang Z, et al. Curr Drug Targets. 2009 Jan;10(1):38-41. doi: 10.2174/138945009787122914. Curr Drug Targets. 2009. PMID: 19149534 Review.
Cited by
- Differential tumorigenic potential and matriptase activation between PDGF B versus PDGF D in prostate cancer.
Najy AJ, Won JJ, Movilla LS, Kim HR. Najy AJ, et al. Mol Cancer Res. 2012 Aug;10(8):1087-97. doi: 10.1158/1541-7786.MCR-12-0071. Epub 2012 Jun 11. Mol Cancer Res. 2012. PMID: 22689130 Free PMC article. - Pharmacological targeting of the PDGF-CC signaling pathway for blood-brain barrier restoration in neurological disorders.
Lewandowski SA, Fredriksson L, Lawrence DA, Eriksson U. Lewandowski SA, et al. Pharmacol Ther. 2016 Nov;167:108-119. doi: 10.1016/j.pharmthera.2016.07.016. Epub 2016 Aug 12. Pharmacol Ther. 2016. PMID: 27524729 Free PMC article. Review. - Insufficiency of hepatocyte growth factor activator inhibitor-1 confers lymphatic invasion of tongue carcinoma cells.
Izumi A, Yamamoto K, Kawaguchi M, Yamashita F, Fukushima T, Kiwaki T, Tanaka H, Yamashita Y, Kataoka H. Izumi A, et al. Cancer Sci. 2022 Jun;113(6):2179-2193. doi: 10.1111/cas.15346. Epub 2022 Apr 12. Cancer Sci. 2022. PMID: 35332604 Free PMC article. - Mechanisms of hepatocyte growth factor activation in cancer tissues.
Kawaguchi M, Kataoka H. Kawaguchi M, et al. Cancers (Basel). 2014 Sep 29;6(4):1890-904. doi: 10.3390/cancers6041890. Cancers (Basel). 2014. PMID: 25268161 Free PMC article. Review. - Changes in neutrophil/lymphocyte and platelet/lymphocyte ratios after chemotherapy correlate with chemotherapy response and prediction of prognosis in patients with unresectable gastric cancer.
Wang F, Liu ZY, Xia YY, Zhou C, Shen XM, Li XL, Han SG, Zheng Y, Mao ZQ, Gong FR, Tao M, Lian L, Li W. Wang F, et al. Oncol Lett. 2015 Dec;10(6):3411-3418. doi: 10.3892/ol.2015.3783. Epub 2015 Oct 8. Oncol Lett. 2015. PMID: 26788143 Free PMC article.
References
- Deuel TF. Polypeptide growth factors: roles in normal and abnormal cell growth. Annu Rev Cell Biol. 1987;3:443–492. - PubMed
- Ross R, Raines EW, Bowen-Pope DF. The biology of platelet-derived growth factor. Cell. 1986;46:155–169. - PubMed
- Rosenkranz S, Kazlauskas A. Evidence for distinct signaling properties and biological responses induced by the PDGF receptor alpha and beta subtypes. Growth Factors. 1999;16:201–216. - PubMed
- Crawford Y, Kasman I, Yu L, Zhong C, Wu X, Modrusan Z, Kaminker J, Ferrara N. PDGF-C mediates the angiogenic and tumorigenic properties of fibroblasts associated with tumors refractory to anti-VEGF treatment. Cancer Cell. 2009;15:21–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32-CA009531/CA/NCI NIH HHS/United States
- R01 CA064139/CA/NCI NIH HHS/United States
- R01 CA123362-01A2/CA/NCI NIH HHS/United States
- T32 CA009531/CA/NCI NIH HHS/United States
- T32 CA009531-21/CA/NCI NIH HHS/United States
- R01 CA123362/CA/NCI NIH HHS/United States
- R29 CA064139/CA/NCI NIH HHS/United States
- F32 CA142038/CA/NCI NIH HHS/United States
- R01 CA123362/CA/NCI NIH HHS/United States
- R01 CA064139-10/CA/NCI NIH HHS/United States
- R01CA064139/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous