Computational modeling and analysis of insulin induced eukaryotic translation initiation - PubMed (original) (raw)
Computational modeling and analysis of insulin induced eukaryotic translation initiation
Joshua Lequieu et al. PLoS Comput Biol. 2011 Nov.
Abstract
Insulin, the primary hormone regulating the level of glucose in the bloodstream, modulates a variety of cellular and enzymatic processes in normal and diseased cells. Insulin signals are processed by a complex network of biochemical interactions which ultimately induce gene expression programs or other processes such as translation initiation. Surprisingly, despite the wealth of literature on insulin signaling, the relative importance of the components linking insulin with translation initiation remains unclear. We addressed this question by developing and interrogating a family of mathematical models of insulin induced translation initiation. The insulin network was modeled using mass-action kinetics within an ordinary differential equation (ODE) framework. A family of model parameters was estimated, starting from an initial best fit parameter set, using 24 experimental data sets taken from literature. The residual between model simulations and each of the experimental constraints were simultaneously minimized using multiobjective optimization. Interrogation of the model population, using sensitivity and robustness analysis, identified an insulin-dependent switch that controlled translation initiation. Our analysis suggested that without insulin, a balance between the pro-initiation activity of the GTP-binding protein Rheb and anti-initiation activity of PTEN controlled basal initiation. On the other hand, in the presence of insulin a combination of PI3K and Rheb activity controlled inducible initiation, where PI3K was only critical in the presence of insulin. Other well known regulatory mechanisms governing insulin action, for example IRS-1 negative feedback, modulated the relative importance of PI3K and Rheb but did not fundamentally change the signal flow.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
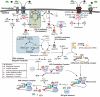
Figure 1. Schematic of the translation initiation signaling network.
Growth factors trigger receptor dimerization and the formation of adaptor complexes which activate PI3K. PI3K then signals through PIP2/3 to activate Akt. Activated Akt can then activate mTORC1 either directly or by phosphorylating TSC1/2, an inhibitor of Rheb. Activated mTORC1 can phosphorylate 4EBP1 and activate S6K1, two necessary checkpoints for translation initiation. mTORC1 can also phosphorylate IRS-1, a negative feedback which inhibits formation of the adaptor complex and attenuates insulin signaling.
Figure 2. The scaled simulation error (SSE) for selected objective function pairs for N = 5818 rank zero initiation models.
The SSEs for objective functions chosen by cross-validation for prediction was set to zero and disregarded when ranking other sets. The red point denotes the performance of the nominal parameter set.
Figure 3. Ensemble performance against selected training objectives (N = 400).
Dotted lines represent the simulation mean of the ensemble, while the shaded region denotes the 99.9% confidence estimate for the mean. The solid dots represent the scaled experimental data. A. Time course data for p70S6K1 phosphorylation in response to insulin stimulation (L6 Myotubes). B. Time course data for c4EBP1 phosphorylation in response to FBS (RhoE 3T3 cells). C. In vitro time course of the 80S complex measured by puromycin assay (rabbit reticulocyte). D. pAkt(Ser473) levels at 20 minutes in the presence and absence of insulin and wortmannin (393T cells). E,F. pAkt(Set473) and activated p70S6K1 levels at 15 minutes in the presence and absence of insulin-like growth factor (IGF) and rapamycin (C2C12 myotubes).
Figure 4. Blind model predictions for the ensemble (N = 400).
The predictive ability of model ensemble was assessed by comparing model performance with novel experimental data. Dotted lines represent the simulation mean of the ensemble, while the shaded region denotes the 99.9% confidence estimate for the mean. The solid dots represent the scaled experimental data. A. In vitro time course for formation of 43S-mRNA complex. A slowly-hydrolyzable GTP homologue (GMP-PNP) was used in place of GTP to isolate formation of this intermediate complex. GMP-PNP data was used for training while GTP data was used for validation. B. Percent of Rheb-GTP to Rheb-GDP in the presence of insulin, wortmannin and rapamycin (A14 NIH 3T3 cells). C. Percent of Rheb-GTP to Rheb-GDP in wildtype and TSC2 lacking cells (MEF cells). D. 4EBP1 bound EIF4E in the presence of heat shock (CHO.K1 cells).
Figure 5. Sensitivity analysis of a population of initiation models (N = 40).
Species with a high sensitivity ranking are considered fragile while species with a low sensitivity ranking are considered robust. A. Sensitivity ranking of network species in the presence and absence of insulin. B. Time-course sensitivity ranking of network species. C,D. Sensitivity ranking of network species in the presence and absence of IRS-1 feedback. Black fill denoted complexes containing IRS-1, grey fill denotes PI3K/Akt associated signaling components. Sensitivity values were time averaged over 0–100 minutes and 0–5 minutes, respectively. Error bars denote one standard error in the sensitivity ranking computed over a family of uncorrelated (mean correlation of approximately 0.6) parameter sets selected for the analysis.
Figure 6. Species knockdown simulations for a population of translation initiation models (N = 400).
Simulated knockdowns were performed by removing nodes from the stoichiometric matrix. The relative change in 80S formation resulting from the removal of a species was used to quantify the impact of the knockdown. A. Species knockdowns in the presence of insulin. Simulated knockdowns resulted in increased (black), constant (white), moderately decreased (dark grey) or severely decreased (light grey) translational levels. B. Species knockouts in the absence of insulin. Simulated knockdowns resulted in increased (black), constant (white), or decreased (grey) translational levels. C. Histogram of translation levels across each member of parameter ensemble. Asterisk index indicates parameter sets that were selected for further analysis. D. Alternative modes of network operation. For a subset of the ensemble, initiation increased following Rheb or mTORC2 disruption. Asterisk indicates rate-limiting step.
Similar articles
- PTEN expression causes feedback upregulation of insulin receptor substrate 2.
Simpson L, Li J, Liaw D, Hennessy I, Oliner J, Christians F, Parsons R. Simpson L, et al. Mol Cell Biol. 2001 Jun;21(12):3947-58. doi: 10.1128/MCB.21.12.3947-3958.2001. Mol Cell Biol. 2001. PMID: 11359902 Free PMC article. - GSK3 and its interactions with the PI3K/AKT/mTOR signalling network.
Hermida MA, Dinesh Kumar J, Leslie NR. Hermida MA, et al. Adv Biol Regul. 2017 Aug;65:5-15. doi: 10.1016/j.jbior.2017.06.003. Epub 2017 Jun 27. Adv Biol Regul. 2017. PMID: 28712664 Review. - Control of oncogenesis by eIF2α phosphorylation: implications in PTEN and PI3K-Akt signaling and tumor treatment.
Koromilas AE, Mounir Z. Koromilas AE, et al. Future Oncol. 2013 Jul;9(7):1005-15. doi: 10.2217/fon.13.49. Future Oncol. 2013. PMID: 23837763 Review.
Cited by
- Multiscale models of breast cancer progression.
Chakrabarti A, Verbridge S, Stroock AD, Fischbach C, Varner JD. Chakrabarti A, et al. Ann Biomed Eng. 2012 Nov;40(11):2488-500. doi: 10.1007/s10439-012-0655-8. Epub 2012 Sep 25. Ann Biomed Eng. 2012. PMID: 23008097 Free PMC article. Review. - Insulin Signaling in Insulin Resistance States and Cancer: A Modeling Analysis.
Bertuzzi A, Conte F, Mingrone G, Papa F, Salinari S, Sinisgalli C. Bertuzzi A, et al. PLoS One. 2016 May 5;11(5):e0154415. doi: 10.1371/journal.pone.0154415. eCollection 2016. PLoS One. 2016. PMID: 27149630 Free PMC article. - Population Heterogeneity in the Epithelial to Mesenchymal Transition Is Controlled by NFAT and Phosphorylated Sp1.
Gould R, Bassen DM, Chakrabarti A, Varner JD, Butcher J. Gould R, et al. PLoS Comput Biol. 2016 Dec 27;12(12):e1005251. doi: 10.1371/journal.pcbi.1005251. eCollection 2016 Dec. PLoS Comput Biol. 2016. PMID: 28027307 Free PMC article. - JuPOETs: a constrained multiobjective optimization approach to estimate biochemical model ensembles in the Julia programming language.
Bassen DM, Vilkhovoy M, Minot M, Butcher JT, Varner JD. Bassen DM, et al. BMC Syst Biol. 2017 Jan 25;11(1):10. doi: 10.1186/s12918-016-0380-2. BMC Syst Biol. 2017. PMID: 28122561 Free PMC article.
References
- Dann SG, Selvaraj A, Thomas G. mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med. 2007;13:252–259. - PubMed
- Sachdev D, Yee D. Disrupting insulin-like growth factor signaling as a potential cancer therapy. Mol Cancer Ther. 2007;6:1–12. - PubMed
- Schneider RJ, Sonenberg N. Translational control in cancer development and progression. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 401–431.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials