Virtual Cell: computational tools for modeling in cell biology - PubMed (original) (raw)
Review
. 2012 Mar-Apr;4(2):129-40.
doi: 10.1002/wsbm.165. Epub 2011 Dec 2.
Affiliations
- PMID: 22139996
- PMCID: PMC3288182
- DOI: 10.1002/wsbm.165
Review
Virtual Cell: computational tools for modeling in cell biology
Diana C Resasco et al. Wiley Interdiscip Rev Syst Biol Med. 2012 Mar-Apr.
Abstract
The Virtual Cell (VCell) is a general computational framework for modeling physicochemical and electrophysiological processes in living cells. Developed by the National Resource for Cell Analysis and Modeling at the University of Connecticut Health Center, it provides automated tools for simulating a wide range of cellular phenomena in space and time, both deterministically and stochastically. These computational tools allow one to couple electrophysiology and reaction kinetics with transport mechanisms, such as diffusion and directed transport, and map them onto spatial domains of various shapes, including irregular three-dimensional geometries derived from experimental images. In this article, we review new robust computational tools recently deployed in VCell for treating spatially resolved models.
Copyright © 2011 Wiley Periodicals, Inc.
Figures
Figure 1
Discretization of diffusion equation on a surface: A local subset of surface grid points is projected on the tangential plane orthogonal to the outward normal vector n at point I (left) and Voronoi tessellation is then applied to projections shown in red (right). This procedure automatically determines the natural neighbors of point i, along with necessary geometric parameters: the sides _s_ij area _A_i of the Voronoi cell and the distances _d_ij between the natural neighbors. In terms of these parameters, the spatially discretized diffusion equation takes the form, Ai∂tUi=∑j∈G(i)D(Uj−Ui)sijsym/dijsym where _U_i are the concentration values at the surface grid points, D is the diffusion coefficient, sijsym=(sij+sij)/2 and dijsym=(dij+dij)/2.
Figure 2
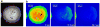
FLIP experiments with fibroblasts expressing GFP-Rac (adapted from (20)). (a) A representative cell, 30 min after replating on fibronectin. The red line delineates the photobleached area. (b) Cell in (a) at the indicated times during the photobleaching protocol.
Figure 3
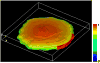
Simulation of a FLIP experiment in VCell using geometry obtained from a z-stack of confocal images (details of how image-based geometries are generated in VCell are given below in section 3). The snapshot represents distribution of the membrane-bound GFP-Rac, in arbitrary units, shortly (0.5 s) after the start of photobleaching. For this time, surface density of the membrane-bound Rac in the unbleached area remains close to maximum.
Figure 4
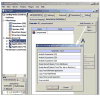
VCell Geometry Editor (VCell Beta 5.0) includes tools for creating geometries both analytically and from experimental images.
Figure 5
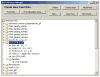
Segmentation of an imported image. The Image Geometry Editor window includes tools for adjusting an uploaded image (cropping (D), magnifying, and adjusting of brightness (C)), as well as tools for segmenting the image ((E), (F), (J), (K), and (H)) necessary for creating a valid VCell geometry.
Figure 6
Field Data Manager display. The Field Data Manager tool allows a user to use irregular spatial distributions from experimental images (or simulation results) as initial conditions in VCell models.
Figure 7
A simple version of the local excitation - global inhibition (LEGI) model (30, 31): (a) simulation geometry and (b) steady-state concentration gradients of the active form of a protein, _P_a, in arbitrary units.
Similar articles
- Virtual Cell modelling and simulation software environment.
Moraru II, Schaff JC, Slepchenko BM, Blinov ML, Morgan F, Lakshminarayana A, Gao F, Li Y, Loew LM. Moraru II, et al. IET Syst Biol. 2008 Sep;2(5):352-62. doi: 10.1049/iet-syb:20080102. IET Syst Biol. 2008. PMID: 19045830 Free PMC article. - The virtual cell.
Schaff J, Loew LM. Schaff J, et al. Pac Symp Biocomput. 1999:228-39. doi: 10.1142/9789814447300_0023. Pac Symp Biocomput. 1999. PMID: 10380200 - Use of virtual cell in studies of cellular dynamics.
Slepchenko BM, Loew LM. Slepchenko BM, et al. Int Rev Cell Mol Biol. 2010;283:1-56. doi: 10.1016/S1937-6448(10)83001-1. Int Rev Cell Mol Biol. 2010. PMID: 20801417 Free PMC article. Review. - Rule-based modeling with Virtual Cell.
Schaff JC, Vasilescu D, Moraru II, Loew LM, Blinov ML. Schaff JC, et al. Bioinformatics. 2016 Sep 15;32(18):2880-2. doi: 10.1093/bioinformatics/btw353. Epub 2016 Jun 9. Bioinformatics. 2016. PMID: 27497444 Free PMC article. - The Virtual Cell: a software environment for computational cell biology.
Loew LM, Schaff JC. Loew LM, et al. Trends Biotechnol. 2001 Oct;19(10):401-6. doi: 10.1016/S0167-7799(01)01740-1. Trends Biotechnol. 2001. PMID: 11587765 Review.
Cited by
- A cell-centered, agent-based framework that enables flexible environment granularities.
Kennedy RC, Ropella GE, Hunt CA. Kennedy RC, et al. Theor Biol Med Model. 2016 Feb 2;13:4. doi: 10.1186/s12976-016-0030-9. Theor Biol Med Model. 2016. PMID: 26839017 Free PMC article. - CellOrganizer: Learning and Using Cell Geometries for Spatial Cell Simulations.
Majarian TD, Cao-Berg I, Ruan X, Murphy RF. Majarian TD, et al. Methods Mol Biol. 2019;1945:251-264. doi: 10.1007/978-1-4939-9102-0_11. Methods Mol Biol. 2019. PMID: 30945250 Free PMC article. - A Modular Workflow for Model Building, Analysis, and Parameter Estimation in Systems Biology and Neuroscience.
Santos JPG, Pajo K, Trpevski D, Stepaniuk A, Eriksson O, Nair AG, Keller D, Hellgren Kotaleski J, Kramer A. Santos JPG, et al. Neuroinformatics. 2022 Jan;20(1):241-259. doi: 10.1007/s12021-021-09546-3. Epub 2021 Oct 28. Neuroinformatics. 2022. PMID: 34709562 Free PMC article. - A conservative algorithm for parabolic problems in domains with moving boundaries.
Novak IL, Slepchenko BM. Novak IL, et al. J Comput Phys. 2014 Aug 1;270:203-213. doi: 10.1016/j.jcp.2014.03.014. J Comput Phys. 2014. PMID: 25067852 Free PMC article. - Numerical Approach to Spatial Deterministic-Stochastic Models Arising in Cell Biology.
Schaff JC, Gao F, Li Y, Novak IL, Slepchenko BM. Schaff JC, et al. PLoS Comput Biol. 2016 Dec 13;12(12):e1005236. doi: 10.1371/journal.pcbi.1005236. eCollection 2016 Dec. PLoS Comput Biol. 2016. PMID: 27959915 Free PMC article.
References
- Loew LM, Schaff JC. The Virtual Cell: a software environment for computational cell biology. Trends Biotechnol. 2001;19:401–6. - PubMed
- Moraru II, Schaff JC, Slepchenko BM, Loew LM. The virtual cell: an integrated modeling environment for experimental and computational cell biology. Ann N Y Acad Sci. 2002;971:595–6. - PubMed
- Slepchenko BM, Schaff JC, Carson JH, Loew LM. Computational cell biology: spatiotemporal simulation of cellular events. Annu Rev Biophys Biomol Struct. 2002;31:423–41. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources