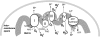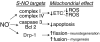Redox regulation of mitochondrial function - PubMed (original) (raw)
Review
Redox regulation of mitochondrial function
Diane E Handy et al. Antioxid Redox Signal. 2012.
Abstract
Redox-dependent processes influence most cellular functions, such as differentiation, proliferation, and apoptosis. Mitochondria are at the center of these processes, as mitochondria both generate reactive oxygen species (ROS) that drive redox-sensitive events and respond to ROS-mediated changes in the cellular redox state. In this review, we examine the regulation of cellular ROS, their modes of production and removal, and the redox-sensitive targets that are modified by their flux. In particular, we focus on the actions of redox-sensitive targets that alter mitochondrial function and the role of these redox modifications on metabolism, mitochondrial biogenesis, receptor-mediated signaling, and apoptotic pathways. We also consider the role of mitochondria in modulating these pathways, and discuss how redox-dependent events may contribute to pathobiology by altering mitochondrial function.
Figures
FIG. 1.
Mitochondrial sites of superoxide production. Mitochondrial electron transport complexes (I–IV) are located on the inner mitochondrial membrane. The electron transport in mitochondria starts with the extraction of electrons from NADH (complex I) or FADH2 (complex II) generated in the tricarboxylic acid (TCA) cycle. Electrons are transferred along the path shown in the figure, resulting in the reduction of O2 to water at complex IV. During this process, protons (H+) are pumped by complexes I, III, and IV into the intermembrane space to form an electrochemical gradient (H+ potential or Δψ) at the inner mitochondrial membrane. ATP synthase utilizes the stored energy of this proton gradient to drive the formation of ATP (proton motive force) from ADP and inorganic phosphate (Pi). Electron leak during respiration leads to the formation of superoxide anion () due to the incomplete reduction of O2 at complex I and complex III (223, 349).
formed at complex I is released into the mitochondrial matrix, whereas
formed at complex III can be released either to the matrix or to the intermembrane space, depending on the disposition of complex III from which it forms.
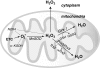
FIG. 2.
Mitochondrial hydrogen peroxide (H2O2) flux. Most of the reactive oxygen species (ROS) generated in mitochondria is in the form of from the electron transport chain (ETC) (223, 349). Recent evidence suggests that NADPH-dependent oxidase 4 (NOX4) may be localized to mitochondria, where it may produce hydrogen peroxide and/or
(118). The TCA cycle enzyme α-ketoglutarate dehydrogenase (α-KGDH) is also a potential source of
(423). Within the mitochondrial matrix, manganese-dependent superoxide dismutase (MnSOD) is the only SOD present (236); it reduces
to hydrogen peroxide. Mitochondrially located glutathione peroxidase-1 (GPx-1) utilizes glutathione (reduced glutathione [GSH]) as a cosubstrate in the reduction of hydrogen peroxide to water (126, 133). Similarly, the mitochondrially targeted peroxiredoxins (Prx3 or Prx5) reduce hydrogen peroxide using thioredoxin 2 (Trx2) to regenerate the active site (66, 73). Also important are glutathione reductase (GR) and Trx reductase (TrxR), which serve to reduce oxidized GSH and Trx, respectively (not represented in this figure). In the intermembrane space, it is thought that copper/zinc-dependent SOD (Cu/ZnSOD) may play a role in reducing
that is released from complex III. Within this space and in the cytosol, GPx-1 and Prx family members reduce hydrogen peroxide produced and released from the mitochondria or other sources.
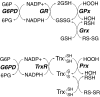
FIG. 3.
Role of NADPH and glucose-6-phosphate dehydrogenase (G6PD) in redox homeostasis. Cellular stores of NADPH are essential for the reductive recycling of GSH and Trx, as it is a necessary cofactor for the enzymes GR and TrxR, which restore the reduced pools of these redox-agents, respectively. [Through different mechanisms involving enzyme stability, NADPH is also essential for the activity of catalase (166, 216)]. As illustrated in this diagram, GSH is necessary for both the activity of GPxs, which reduce hydrogen and lipid peroxides (133), and glutaredoxin (Grx), which reduces protein-mixed disulfides with GSH (RS-SG) (209). To some extent, Grx may also reduce protein disulfides as well (not in the diagram), similar to the actions of Trx, which plays an important role in modulating protein disulfides, including those of peroxiredoxins (Prx). Trx is, thus, essential for Prx-mediated hydrogen peroxide reduction. In mitochondria, GPx-1 and GPx-4 modulate the reduction of hydrogen and lipid peroxides. Mitochondrially localized GR and Grx2, as well as their cytoplasmic forms, contribute to overall GSH/oxidized glutathione (GSSG) redox status. Prx3 and Prx5, along with Trx2 and TrxR2, comprise the mitochondrial Trx system. Extracellular and cytosolic redox potential may also influence the redox status of these antioxidant proteins.
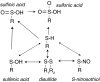
FIG. 4.
Thiol oxidation. Protein cysteinyl residues form a large redox-active thiol pool, the modifications of which can regulate protein structure and function (197, 463, 464). Protein cysteines (Cys) that exist as thiolates (S−) rather than as S-H may be especially susceptible to modification (not represented in the figure) (368). Oxidative stress can stimulate reversible modification after sulfenic acid or disulfide bond formation. Disulfides can form intramolecularly, between proteins or between a protein and a low-molecular-weight redox active thiol, such as GSH. Trx and Grx may play a role in reducing oxidant-stimulated (reversible) oxidations. Excess oxidant stress is associated with the overoxidation of protein thiols to sulfinic acid or sulfonic acid. These thiol oxidation states are considered irreversible, except for the special case of peroxiredoxins: sulfiredoxin specifically targets and reduces sulfinic acids in some peroxiredoxins (74).
FIG. 5.
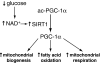
Glucose metabolism and NADH/NAD+. The enzymes, pyruvate dehydrogenase (PDH) and lactate dehydrogenase (LDH), dictate whether glycolysis proceeds to glucose oxidation or the formation of lactate. PDH can be inhibited by fatty acid oxidation (341), c-jun N-terminal kinase (JNK) signaling (480), and hypoxia-mediated upregulation of PDH kinase, which, similar to JNK, phosphorylates and inactivates PDH (386). LDH activity is dependent on the expression of the lactate dehydrogenase A (LDHA) or lactate dehydrogenase B (LDHB) subtypes, which promote either lactate formation or pyruvate formation, respectively. The absence of glucose or other conditions that elevate NAD+ promotes sirtuin (SIRT) deacetylase activity to stimulate respiration, activate glycolytic genes, and increase fatty acid oxidation (219) (not shown in figure).
FIG. 6.
Regulation of metabolism by SIRT1. SIRT can affect mitochondrial function in several ways. SIRT1 has been shown to modulate mitochondrial biogenesis and fatty acid oxidation (62, 354). A decrease in cellular glucose leads to an increase in intracellular NAD+ to promote SIRT1 deacetylase activity. Peroxisome proliferator-activated receptor-γ coactivator-1 alpha (PGC-1α) is one of the targets of SIRT1 (33). Deacetylation of this transcriptional coactivator promotes the biogenic program (in part, via nuclear respiratory factors, not represented in this diagram) to increase cellular mitochondrial content (150). In addition, in highly metabolic cells, such as skeletal muscle, PGC-1α activation increases the expression of genes involved in fatty acid oxidation to increase mitochondrial respiration (150). Activation of other SIRTs, such as SIRT3, may also contribute to these pathways (not shown in the diagram). SIRT3 targets to mitochondria, and its deacetylation of mitochondrial proteins may augment complex I activity (6, 389) and increase fatty acid oxidation by maintaining the activation of the long-chain acyl coenzyme A dehydrogenase, at least in liver (175).
FIG. 7.
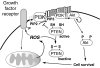
S-NO targets and mitochondrial function. Nitric oxide (NO·) can mediate protein S-nitrosation (-S-NO) or activate soluble guanylyl cyclase to foster cGMP-mediated kinase function. Protein –S-NO has been shown to occur in response to physiological and pathological production of NO· (463), and NO· has been shown to regulate mitochondrial function through several mechanisms. Complex I S-nitrosation has been associated with diminished activity; however, conflicting results exist as to whether it inhibits or promotes mitochondrial ROS generation (57, 98, 108). Similarly, S-nitrosation of complex IV has been suggested to inhibit mitochondrial respiration (97, 474). In general, low concentrations of NO· are associated with antiapoptotic mechanisms, perhaps, in part, via the inhibition of caspase 3 activation or the decreased degradation of the antiapoptotic Bcl-2 following their respective S-nitrosation (21, 424). Nitric oxide may enhance cell death and mitochondrial fragmentation in neurodegenerative diseases by its effects on dynamin-related protein-1 (Drp1), which promotes mitochondrial fission (86). There is some controversy, however, as to whether these are cGMP-kinase-mediated or –S-NO-specific effects (45). In addition, in myogenesis, NO· has the opposite effect on mitochondrial fission, promoting the fusion of mitochondria into an elongated network by inhibiting Drp1-mediated fission (113).
FIG. 8.
The role of mitochondria in growth factor-mediated signaling. Growth factor receptor signaling promotes mitochondrial ROS, without which signaling responses are diminished (169). Mitochondrial ROS serve to activate receptor phosphorylation (402) and promote intracellular signaling, in part, by the oxidative inactivation of phosphatases (114). Illustrated in this figure is the role of ROS in Akt signaling, a process in which mitochondrial ROS are essential (269). Phosphatidylinositol 3 kinase (PI3K) phosphorylates membrane phosphatidylinositol-4,5-phosphate (PIP2) to PIP3 (3,4,5 phosphate). Accumulation of PIP3 in the membrane recruits PDH kinase 1 (PDK1) and Akt to the membrane, where PDK1 phosphorylates Akt (269). Akt may also be phosphorylated by other kinases, or self-phosphorylate to a certain degree. Activated Akt targets other cellular proteins to promote cell survival and may also enhance mitochondrial biogenesis. The actions of PI3K are antagonized by the lipid phosphatase phosphatase and tensin homolog (PTEN), which dephosphorylates PIP3. PTEN is inactivated by redox modification (330): in the presence of hydrogen peroxide, PTEN forms an intramolecular disulfide bond. Oxidative inactivation of PTEN promotes growth factor signaling. Similarly, oxidative inactivation of other phosphatases, such as PTP1B or MAP kinase phosphatases, is necessary for essential signal transduction downstream of insulin-mediated receptor activation or other signaling events, respectively.
FIG. 9.
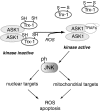
Redox regulation of JNK activation and apoptotic pathways. JNK is one of the stress-activated protein kinases (SAPK) (230). Apoptosis signal-regulated kinase 1 (ASK1) is the mitogen-activated protein kinase (MAPK) kinase that regulates the SAPK response to oxidative stimuli. The redox regulation of ASK1 is indirect and due to protein–protein interactions with Trx1 (365). Reduced Trx1 association with ASK maintains ASK in its inactive form (141). Oxidation of Trx1 dissociates it from ASK, leading to the association of ASK with other proteins, including the tumor necrosis factor-receptor-associated adaptor proteins (TRAFs), which may promote ASK activation (282, 283). Activated ASK is free to target JNK, which, once phosphorylated, can promote a pro-apoptotic response. JNK phosphorylation of downstream nuclear factors can increase the expression of pro-apoptotic genes, and its phosphorylation of mitochondrial targets, such as p66shc, can enhance mitochondrial ROS production and apoptogen release.
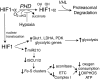
FIG. 10.
Oxygen tension, hypoxia-inducible factor-1 (HIF-1) activation of miR210 and mitochondrial function. Under normal O2 tension, HIF-1α is hydroxylated by the prolyl hydroxylases (PHD), which utilize O2 and α-ketoglutarate (αKG) as cosubstrates (203). Hydroxylated HIF-1α is targeted for ubiquitination after recognition by the von Hippel-Lindau tumor suppressor (VHL) protein, leading to its subsequent proteasomal degradation. Under hypoxia, decreased O2 tension suppresses PHD activity, allowing for HIF-1α translocation to the nucleus, where it combines with HIF-β, which is not sensitive to O2 tension, to form the transcriptionally active dimer HIF-1. HIF-1 upregulates gene expression of the glucose transporter-1 (Glut1), LDHA, PDK, and glycolytic genes to promote an increase in glycolysis (385, 386). HIF-1 also transcriptionally activates the expression of miR210, which inhibits translation of iron–sulfur (Fe-S) cluster assembly protein isoforms ISCU1 and ISCU2 (69). A loss of Fe-S cluster formation results in a suppression of ETC complexes, including complex I and aconitase, which rely on Fe-S prosthetic groups for their function. The consequences of diminished Fe-S cluster formation suppress ROS in hypoxia, reduce oxygen consumption, decrease respiration, and decrease oxidative phosphorylation (Pasteur effect) (69).
FIG. 11.
ROS, redox, and mitochondria. Growth factors, nutrients, Ca2+ fluxes, ROS and its generators (including angiotensin II [AII] and carbon monoxide [CO]), and NO· modulate mitochondrial function in a number of ways, including modulating respiration, promoting mitochondrial ROS production, or regulating cellular mitochondrial content by stimulating biogenic or mitophagy mediators. Cellular antioxidants and uncoupling proteins (UCP) modulate cellular responses to essential and excess cellular ROS. Mitochondria both generate and respond to these ROS. Essential ROS may be generated during normal respiration or in response to growth factor signaling, which stimulates ROS generation from mitochondria. Lack of this essential signaling can alter cellular redox balance, protein disulfide formation, and cell signaling via reversible thiol oxidation (for instance, of PTEN or DUSP phosphatases) or other mechanisms (169, 264, 464). Antioxidants like GPx-1 may attenuate these responses. Essential ROS may also play a role in modulating mitochondrial biogenesis (350). AII receptor activation is one stimulus that may lead to the generation of excess ROS. Thus, AII-mediated stimulation of NOX-dependent ROS was found to enhance mitochondrial production of ROS to promote mitochondrial dysfunction (121). Excess ROS can cause oxidative damage to members of the ETC, such as complex I, or members of the TCA cycle, such as aconitase or α-ketoglutarate, or other mitochondrial proteins (4). Excess ROS can also promote the mitochondrial recruitment of proteins such as p66shc that further amplify ROS production, thereby contributing to cell death (ROS-mediated ROS generation) (297). Other mitochondrial proteins, such as mitochondrial STAT3 (mSTAT3), appear to preserve ETC activity by preventing ROS leak at complex I. Excess ROS leads to redox imbalance, overoxidation of protein thiols, oxidation of mitochondrial lipids, and mitochondrial permeability transition (MPT). At some threshold of ROS, these changes result in necrosis or apoptosis. Cells possess several mechanisms to prevent excess ROS, including the biogenic program, which generates new mitochondria and increases the expression of some antioxidant genes (398), as well as mitophagy, which may target and remove damaged mitochondria (469). ROS upregulation of antioxidants or UCP may also modulate the responses of excess ROS (301), and, under some circumstances, brief MPT may alleviate mitochondrial stress to prevent mitochondrial dysfunction (123).
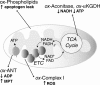
FIG. 12.
Mitochondrial targets oxidatively modified by ROS. ROS can directly modulate the mitochondria by reacting with many redox-sensitive targets. Among these targets, membrane lipid peroxidation (ox-phospholipids) can lead to the release of apoptogens, such as cytochrome c and apoptosis-inducing factor, causing subsequent apoptosis. Antioxidant enzymes, such as GPx-4, which can reduce oxidized membrane phospholipids, attenuate these apoptotic modifications (316, 340). Enzymes in the TCA cycle, such as aconitase or α-KGDH, are susceptible to oxidative inactivation (ox-) (423). Decreased flux through the TCA cycle results in decreased NADH production, thereby reducing electron transport (ETC) and oxidative phosphorylation. Oxidation of the adenine nucleotide transporter (ANT) results in the formation of intramolecular disulfide bonds that alters the ability of the transporter to uptake ADP (101). This inactivation of the ANT sensitizes mitochondria to a permeability transition (MPT), leading to the loss of integrity of the mitochondrial membrane (potential), mitochondrial swelling, and cell death. Although other ETC complexes are also susceptible to oxidative modification, oxidation of complex I is associated with decreased complex I activity and increased production of superoxide (120, 416).
Similar articles
- Redox regulation of mitochondrial function with emphasis on cysteine oxidation reactions.
Mailloux RJ, Jin X, Willmore WG. Mailloux RJ, et al. Redox Biol. 2013 Dec 19;2:123-39. doi: 10.1016/j.redox.2013.12.011. eCollection 2014. Redox Biol. 2013. PMID: 24455476 Free PMC article. Review. - Mitochondrial redox signaling during apoptosis.
Cai J, Jones DP. Cai J, et al. J Bioenerg Biomembr. 1999 Aug;31(4):327-34. doi: 10.1023/a:1005423818280. J Bioenerg Biomembr. 1999. PMID: 10665523 Review. - Protein S-glutathionlyation links energy metabolism to redox signaling in mitochondria.
Mailloux RJ, Treberg JR. Mailloux RJ, et al. Redox Biol. 2016 Aug;8:110-8. doi: 10.1016/j.redox.2015.12.010. Epub 2015 Dec 31. Redox Biol. 2016. PMID: 26773874 Free PMC article. Review. - Cellular metabolic and autophagic pathways: traffic control by redox signaling.
Dodson M, Darley-Usmar V, Zhang J. Dodson M, et al. Free Radic Biol Med. 2013 Oct;63:207-21. doi: 10.1016/j.freeradbiomed.2013.05.014. Epub 2013 May 20. Free Radic Biol Med. 2013. PMID: 23702245 Free PMC article. Review. - Apoptotic interactions of cytochrome c: redox flirting with anionic phospholipids within and outside of mitochondria.
Bayir H, Fadeel B, Palladino MJ, Witasp E, Kurnikov IV, Tyurina YY, Tyurin VA, Amoscato AA, Jiang J, Kochanek PM, DeKosky ST, Greenberger JS, Shvedova AA, Kagan VE. Bayir H, et al. Biochim Biophys Acta. 2006 May-Jun;1757(5-6):648-59. doi: 10.1016/j.bbabio.2006.03.002. Epub 2006 Mar 31. Biochim Biophys Acta. 2006. PMID: 16740248 Review.
Cited by
- Antioxidant functions of DHHC3 suppress anti-cancer drug activities.
Sharma C, Yang W, Steen H, Freeman MR, Hemler ME. Sharma C, et al. Cell Mol Life Sci. 2021 Mar;78(5):2341-2353. doi: 10.1007/s00018-020-03635-3. Epub 2020 Sep 28. Cell Mol Life Sci. 2021. PMID: 32986127 Free PMC article. - Physiological Concentration of Exogenous Lactate Reduces Antimycin A Triggered Oxidative Stress in Intestinal Epithelial Cell Line IPEC-1 and IPEC-J2 In Vitro.
Kahlert S, Junnikkala S, Renner L, Hynönen U, Hartig R, Nossol C, Barta-Böszörményi A, Dänicke S, Souffrant WB, Palva A, Rothkötter HJ, Kluess J. Kahlert S, et al. PLoS One. 2016 Apr 7;11(4):e0153135. doi: 10.1371/journal.pone.0153135. eCollection 2016. PLoS One. 2016. PMID: 27054581 Free PMC article. - Mitochondrial Porin Is Involved in Development, Virulence, and Autophagy in Fusarium graminearum.
Han X, Li Q, Li X, Lv X, Zhang L, Zou S, Yu J, Dong H, Chen L, Liang Y. Han X, et al. J Fungi (Basel). 2022 Sep 4;8(9):936. doi: 10.3390/jof8090936. J Fungi (Basel). 2022. PMID: 36135661 Free PMC article. - Efficacy of Nutritional Interventions as Stand-Alone or Synergistic Treatments with Exercise for the Management of Sarcopenia.
Damanti S, Azzolino D, Roncaglione C, Arosio B, Rossi P, Cesari M. Damanti S, et al. Nutrients. 2019 Aug 23;11(9):1991. doi: 10.3390/nu11091991. Nutrients. 2019. PMID: 31443594 Free PMC article. Review. - Mannose inhibits the growth of prostate cancer through a mitochondrial mechanism.
Deng YL, Liu R, Cai ZD, Han ZD, Feng YF, Cai SH, Chen QB, Zhu JG, Zhong WD. Deng YL, et al. Asian J Androl. 2022 Sep-Oct;24(5):540-548. doi: 10.4103/aja2021104. Asian J Androl. 2022. PMID: 35142655 Free PMC article.
References
- Abu-Soud HM. Yoho LL. Stuehr DJ. Calmodulin controls neuronal nitric-oxide synthase by a dual mechanism. Activation of intra- and interdomain electron transfer. J Biol Chem. 1994;269:32047–32050. - PubMed
- Adachi T. Pimentel DR. Heibeck T. Hou X. Lee YJ. Jiang B. Ido Y. Cohen RA. S-glutathiolation of Ras mediates redox-sensitive signaling by angiotensin II in vascular smooth muscle cells. J Biol Chem. 2004;279:29857–29862. - PubMed
- Agostinelli E. Marques MP. Calheiros R. Gil FP. Tempera G. Viceconte N. Battaglia V. Grancara S. Toninello A. Polyamines: fundamental characters in chemistry and biology. Amino Acids. 2010;38:393–403. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 HL108630/HL/NHLBI NIH HHS/United States
- U54 HL070819/HL/NHLBI NIH HHS/United States
- P01 HL048743/HL/NHLBI NIH HHS/United States
- R01 HL061795/HL/NHLBI NIH HHS/United States
- HL108630/HL/NHLBI NIH HHS/United States
- HL48743/HL/NHLBI NIH HHS/United States
- HL107192/HL/NHLBI NIH HHS/United States
- HL70819/HL/NHLBI NIH HHS/United States
- HL61795/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources