Role of DNMT3B in the regulation of early neural and neural crest specifiers - PubMed (original) (raw)
Role of DNMT3B in the regulation of early neural and neural crest specifiers
Kristen Martins-Taylor et al. Epigenetics. 2012.
Abstract
The de novo DNA methyltransferase DNMT3B functions in establishing DNA methylation patterns during development. DNMT3B missense mutations cause immunodeficiency, centromere instability and facial anomalies (ICF) syndrome. The restriction of Dnmt3b expression to neural progenitor cells, as well as the mild cognitive defects observed in ICF patients, suggests that DNMT3B may play an important role in early neurogenesis. We performed RNAi knockdown of DNMT3B in human embryonic stem cells (hESCs) in order to investigate the mechanistic contribution of DNMT3B to DNA methylation and early neuronal differentiation. While DNMT3B was not required for early neuroepithelium specification, DNMT3B deficient neuroepithelium exhibited accelerated maturation with earlier expression, relative to normal hESCs, of mature neuronal markers (such as NEUROD1) and of early neuronal regional specifiers (such as those for the neural crest). Genome-wide analyses of DNA methylation by MethylC-seq identified novel regions of hypomethylation in the DNMT3B knockdowns along the X chromosome as well as pericentromeric regions, rather than changes to promoters of specific dysregulated genes. We observed a loss of H3K27me3 and the polycomb complex protein EZH2 at the promoters of early neural and neural crest specifier genes during differentiation of DNMT3B knockdown but not normal hESCs. Our results indicate that DNMT3B mediates large-scale methylation patterns in hESCs and that DNMT3B deficiency in the cells alters the timing of their neuronal differentiation and maturation.
Figures
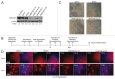
Figure 1. Early NE specification is not affected by DNMT3B knockdown. (A) protein gel blot analyses of DNMT3B to confirm DNMT3B knockdown efficiency in hESCs. β-actin was used as a loading control. For each lane, the DNMT3B/ β-actin ratio is shown, as determined by quantification of the bands. The relative amount of remaining DNMT3B in the KDs was determined relative to undifferentiated H9 hESCs. (B) Schematic of NE differentiation protocol. Morphogens can be added during differentiation as follows: * = Addition of exogenous morphogens, such as retinoic acid, at day 10 (d10) of differentiation is required for robust expression of neural patterning genes around d17. ** = Addition of exogenous morphogens to hESC-derived rosettes beginning at d16 is required for robust expression of NC patterning genes at d28. (C) Phase contrast images for NE at d10. (D) Immunocytochemistry for PAX6 and TUJ1 at d10. Cell nuclei were counterstained with DAPI.
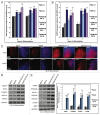
Figure 3. DNMT3B knockdown enhances NC lineage differentiation and specification. (A, B) Gene expression analyses of PAX7 (A), or NGFR (B) at d5 and d7 of differentiation. Data was normalized to undifferentiated H9 hESCs and displayed as a Log2 ratio. (C) Immunocytochemistry for PAX7 and NGFR at d8. Cell nuclei were counterstained with DAPI. (D, E) Gene expression analyses of early NC specifiers at d10 (D), or NC lineage fate determinants (E) at d12. Gene expression was also quantified for the NC lineage fate determinants ASCL1, PHOX2B, HOXC8, and S100β at d12 of differentiation. Data was normalized to undifferentiated H9 hESCs and displayed as a Log2 ratio. The p values displayed correspond as follows: * < 0.10, ** < 0.05, *** < 0.01.
Figure 2. Expression of neural genes that specify regional identity is altered when DNMT3B is knocked down. (A) Gene expression analyses of NE regional specifiers at d12 of differentiation. Brain regions specified by these genes are indicated. (B) Immunocytochemistry for HOXB4 and ISL1 at d12. Cell nuclei were counterstained with DAPI. (C) Gene expression quantification of EN1, TH, HOXB4, ISL1 at d0 (top) and d12 (bottom) of differentiation. Data was normalized to undifferentiated H9 hESCs and displayed as a Log2 ratio. The p values displayed correspond as follows: * = < 0.05.
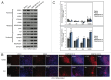
Figure 4. Aberrant DNA methylation in DNMT3B KDs. (A) Chromosome-wide distribution of DNA methylation along X chromosome. HMDs with > 80% DNA methylation (common to all samples) are marked in blue. (B) The average percent methylation observed within HMDs and PMDs along X chromosome. Control samples used include male H1 hESCs and male cerebral cortex, which showed no differences in the methylation of these two domains. (C) The average percent methylation of autosomal domains. Control sample used include male H1 hESCs. The p values displayed correspond as follows: *** = < 0.001.
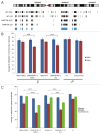
Figure 5. DNMT3B associates with the promoters of early neural determinant genes. (A) DNMT3B association at PAX6, NEUORD1, PAX7, and NGFR promoters in MIS24 and DNMT3B KD 13 hESCs. DNMT3B was also enriched at known target promoters of ID2 and HHEX, but not at GAPDH. Quantification of ChIP was performed and displayed as % input for each sample. (B) Promoter regions are “bivalent” in hESCs, with H3K27me3 and H3K4me3. (C) Association of EZH2 at the promoters in hESCs. (D) Levels of H3K27me3 and EZH2 at d7 of differentiation. Quantification of ChIP was performed and displayed as % input for each sample. The p values displayed correspond as follows: * = < 0.08, ** = < 0.05, *** = < 0.01.
Similar articles
- DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development.
Okano M, Bell DW, Haber DA, Li E. Okano M, et al. Cell. 1999 Oct 29;99(3):247-57. doi: 10.1016/s0092-8674(00)81656-6. Cell. 1999. PMID: 10555141 - Involvement of DNMT3B in the pathogenesis of Hirschsprung disease and its possible role as a regulator of neurogenesis in the human enteric nervous system.
Torroglosa A, Enguix-Riego MV, Fernández RM, Román-Rodriguez FJ, Moya-Jiménez MJ, de Agustín JC, Antiñolo G, Borrego S. Torroglosa A, et al. Genet Med. 2014 Sep;16(9):703-10. doi: 10.1038/gim.2014.17. Epub 2014 Feb 27. Genet Med. 2014. PMID: 24577265 - The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease.
Ehrlich M. Ehrlich M. Clin Immunol. 2003 Oct;109(1):17-28. doi: 10.1016/s1521-6616(03)00201-8. Clin Immunol. 2003. PMID: 14585272 Review. - DNA methylation in disease: Immunodeficiency, Centromeric instability, Facial anomalies syndrome.
Vukic M, Daxinger L. Vukic M, et al. Essays Biochem. 2019 Dec 20;63(6):773-783. doi: 10.1042/EBC20190035. Essays Biochem. 2019. PMID: 31724723 Free PMC article. Review.
Cited by
- Whole-genome bisulfite DNA sequencing of a DNMT3B mutant patient.
Heyn H, Vidal E, Sayols S, Sanchez-Mut JV, Moran S, Medina I, Sandoval J, Simó-Riudalbas L, Szczesna K, Huertas D, Gatto S, Matarazzo MR, Dopazo J, Esteller M. Heyn H, et al. Epigenetics. 2012 Jun 1;7(6):542-50. doi: 10.4161/epi.20523. Epub 2012 Jun 1. Epigenetics. 2012. PMID: 22595875 Free PMC article. - Epigenetic control of gene regulation during development and disease: A view from the retina.
Corso-Díaz X, Jaeger C, Chaitankar V, Swaroop A. Corso-Díaz X, et al. Prog Retin Eye Res. 2018 Jul;65:1-27. doi: 10.1016/j.preteyeres.2018.03.002. Epub 2018 Mar 12. Prog Retin Eye Res. 2018. PMID: 29544768 Free PMC article. Review. - Reactivation of maternal SNORD116 cluster via SETDB1 knockdown in Prader-Willi syndrome iPSCs.
Cruvinel E, Budinetz T, Germain N, Chamberlain S, Lalande M, Martins-Taylor K. Cruvinel E, et al. Hum Mol Genet. 2014 Sep 1;23(17):4674-85. doi: 10.1093/hmg/ddu187. Epub 2014 Apr 23. Hum Mol Genet. 2014. PMID: 24760766 Free PMC article. - Specific ZNF274 binding interference at SNORD116 activates the maternal transcripts in Prader-Willi syndrome neurons.
Langouët M, Gorka D, Orniacki C, Dupont-Thibert CM, Chung MS, Glatt-Deeley HR, Germain N, Crandall LJ, Cotney JL, Stoddard CE, Lalande M, Chamberlain SJ. Langouët M, et al. Hum Mol Genet. 2020 Nov 25;29(19):3285-3295. doi: 10.1093/hmg/ddaa210. Hum Mol Genet. 2020. PMID: 32977341 Free PMC article. - X Chromosome Inactivation Timing is Not e_XACT_: Implications for Autism Spectrum Disorders.
LaSalle JM. LaSalle JM. Front Genet. 2022 Mar 9;13:864848. doi: 10.3389/fgene.2022.864848. eCollection 2022. Front Genet. 2022. PMID: 35356429 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- 2R01HD041462/HD/NICHD NIH HHS/United States
- R01 ES021707/ES/NIEHS NIH HHS/United States
- R01 NS081913/NS/NINDS NIH HHS/United States
- R01 HD048799/HD/NICHD NIH HHS/United States
- R01 HD041462/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases