Ubiquitination, localization, and stability of an anti-apoptotic BCL2-like protein, BCL2L10/BCLb, are regulated by Ubiquilin1 - PubMed (original) (raw)
Ubiquitination, localization, and stability of an anti-apoptotic BCL2-like protein, BCL2L10/BCLb, are regulated by Ubiquilin1
Levi J Beverly et al. Proc Natl Acad Sci U S A. 2012.
Abstract
We have previously shown that all six members of the anti-apoptotic BCL2 gene family can cooperate with (myelocytomatosis oncogene) MYC in a mouse model of leukemia, but three of them are significantly less potent contributors to leukemogenicity than the other three. The protein encoded by one of these less potent genes, BCL2L10/BCLb, was recently shown to vary dramatically in many primary human cancers by immunohistochemistry, and the protein levels were inversely correlated with survival in patients with several cancer types. We examined BCLb mRNA in a panel of human cancer cell lines and did not observe the extensive variation in mRNA that would be required to explain the vast differences in protein levels. We found that the levels of BCLb protein diminish quickly after inhibition of protein synthesis with cycloheximide, so we searched for interacting proteins that might affect posttranslational stability of BCLb. Using a variety of approaches, including immunoaffinity and mass spectrometry, we identified a protein, Ubiquilin1 (Ubqln), that specifically interacts with BCLb, and not with other anti-apoptotic BCL2-like proteins. Ubqln stabilizes BCLb protein, while also promoting monoubiquitination on multiple lysine residues and relocation to the cytosol. Furthermore, primary lung adencarcinomas have more Ubqln mRNA than normal adjacent lung tissue, and higher Ubqln mRNA levels are associated with shorter survival of lung cancer patients, suggesting that potentiation of the anti-apoptotic potential of BCLb through regulation of its stability by Ubqln may be an important factor in tumor progression.
Conflict of interest statement
The authors declare no conflict of interest .
Figures
Fig. 1.
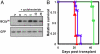
BCLb is a labile protein that is not regulated by alterations in mRNA levels. (A) Invariant levels of BCLb mRNA in cell lines. The mRNA levels for all six anti-apoptotic BCL2 family genes were analyzed in a set of 732 cancer cell lines from diverse tissues of origin using microarray data from the Sanger Institute’s Cancer Cell Line Project (
http://www.sanger.ac.uk/genetics/CGP/CellLines/
). Normalized log2 intensity values for each gene were median-centered across all the cell lines and plotted to show the variation in expression. Box-plots depict the median group expression (mid line), the 25th and 75th percentiles (box), and the limits of 95% of samples for each group “whiskers” with values for all other samples falling outside of this range represented by black dots. (B) Varied response of BCL2 proteins following cycloheximide treatment. 293T cells were transfected with plasmids expressing GFP and BCLb or BCLxl. Forty-eight hours posttransfection cells were left untreated (0) or treated with cycloheximide (+cycloheximide) for the indicated amount of time (in hours). Western blot analysis of lysates was performed with antibodies specific to BCLb or BCLxl. GFP, expressed from the same mRNA as the respective BCL2-proteins, was detected as a loading control. V = empty vector containing only GFP.
Fig. 2.
Lysineless BCLb (BCLbK0) is more stable and a more potent oncogene than wild-type BCLb. (A) Stability of BCLbK0 protein following inhibition of protein synthesis. 293T cells were transfected with BCLbK0 and 48 h posttransfection cells were left untreated (0) or treated with cycloheximide (+cycloheximide) for the indicated amount of time (in hours). BCLb protein levels were determined with an anti-BCLb antibody. Bottom box, western blot analysis for GFP. V = empty vector containing only GFP (B) BCLbK0 is a more potent oncogene. Kaplan-Meier survival analysis of mice reconstituted with cells expressing MYC and BCLbwt (circles), BCLbK0 (crosses) or BCLxl (triangles) (plot is a combination of two independent experiments where nine animals were followed for each group; n = 9).
Fig. 3.
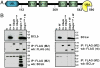
Ubqln interacts specifically with BCLb. (A) Schematic of the Ubquilin1 protein. UBL domain; STI domain; UBA domain. The numbers below the diagram indicate amino acid residues. (B) Ubqln interacts specifically with BCLb. 293T cells were transfected with plasmids containing BCLb (B) and FLAG-tagged Ubqln (left box) or BCLw (W) and FLAG-tagged Ubqln (right box). Forty-eight hours posttransfection cell lysates were prepared and immunoprecipitations were performed with monoclonal anti-FLAG antibodies (M2) (to immunoprecipitate Ubqln-containing complexes). Western blots were performed to detect Ubqln (FLAG rb; poly-clonal FLAG antibody; middle box) and BCLb or BCLw (bottom box). Top box represents 10% of input used for immunoprecipitations. V = empty vector, Ubqln112_X_ = Ubqln encoding the first 112 amino acid of Ubqln containing only the UBL domain. The band migrating at approximately 40 kDa in both the right and left bottom boxes is a nonspecific band detected by the antibodies.
Fig. 4.
Ubqln promotes stabilization of BCLb and accumulation of monoubiquitinated BCLb. (A) BCLb is ubiquitinated on multiple lysine residues. Lysates were prepared from 293T cells expressing BCLb and FLAG-Ubqln, in addition to wild-type HA-ubiquitin (Ubwt), lysineless HA-ubiquitin (UbK0) or HA-ubiquitin-containing only lysine 48 (UbK48). Immunoprecipitations were performed with anti-HA antibodies (to immunoprecipitate Ubiquitin-containing proteins) and followed by Western blots with anti-BCLb antibodies (bottom box). Top three boxes represent 10% of the input used for immunoprecipitations, and anti-Tubulin antibodies were used to show equal amount of total protein extracts were used for immunoprecipitations. (B) Endogenous Ubqln can promote ubiquitination of BCLb. 293T cells were transfected with either nontargeting siRNA (si_N.T.) or siRNA against Ubqln (si_Ubqln). Twenty-four hours later these cells were transfected with vectors encoding BLCb (B) and/or HA-tagged ubiquitin (Ub). Twenty-four hours later cell lysates were prepared and immunoprecipitations were performed with anti-HA antibodies (to immunoprecipitate Ubiquitin-containing proteins) and Western blots to detect BCLb (middle lower box) and total ubiquitinated proteins (lower box) were performed. Top two boxes each represent 10% of the input used for immunoprecipitations. Endogenous Ubqln levels were detected with a poly-clonal antibody raised against Ubqln. The arrow in the middle lower box represents the nonubiquitinated form of BCLb. (C) Ubqln stabilizes BCLb following cycloheximide treatment. Cells expressing BCLb, or BCLb and FLAG-Ubqln were treated with cycloheximide (CH) for 16 h. Cell lysates were prepared and Western blots were performed using anti-BCLb antibodies (top box), anti-FLAG antibodies to detect Ubqln (middle box) or anti-GFP antibodies as controls (bottom box).
Fig. 5.
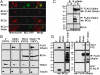
Ubqln alters subcellular localization of BCLb, and preferentially interacts with BCLb in the cytoplasm. (A) Ubqln alters the subcellular localization of BCLb, but not BCL2. H358 cells expressing GFP-tagged BCLb or GFP-tagged BCL2 and either empty vector or a vector containing Ubqln were treated with Mito-Tracker dye and subjected to confocal microscopy. (B) Ubqln promotes the accumulation of ubiquitinated BCLb in the cytosol. 293T cells expressing BCLb, Ubqln, or BCLb and Ubqln were subjected to subcellular fractionation by differential centrifugation, as described in Materials and Methods, followed by Western blots. Cells were fractionated into n-nuclear, m-membranes, c-cytosolic, and s-structural/cytoskeletal compartments. Western blots of VDAC1, a membrane bound protein, and tubulin, a cytoplasmic protein, anti-sera were used to document the validity of the fractionations. (C) Ubqln and BCLb interact in the cytosol. Fractionated lysates expressing either BCLb or BCLb and Ubqln were subjected to immunoprecipitation with anti-FLAG antibody to detect Ubqln interactions. Only the cytosolic (c) and membranous (m) fractions were used, as these were the only fractions previously determined to contain BCLb (B) or Ubqln. Following anti-FLAG immunoprecipitations, Western blots were performed to detect Ubqln and BCLb. BCLb was also detected using 10% of the total protein used for immunoprecipitations (bottom box). (D) Ubiquitinated BCLb interacts preferentially with Ubqln. Cell extracts containing BCLb, FLAG-Ubqln, or both BCLb and FLAG-Ubqln were used to immunoprecipitate Ubqln using anti-FLAG antibody. Following immunoprecipitation, Western blots were performed with both the precipitate (IP: FLAG) and the remaining supernatant (post FLAG IP). Antibodies against BCLb and against FLAG (to detect Ubqln) were used to probe the Western blots.
Fig. 6.
Levels of Ubqln RNA in human lung adenocarcinoma predict survival outcome in patients. (A) Ubqln RNA was analyzed in a set of 46 tumor/normal pairs from the same patients. The microarray platform contained two individual probes for each gene that are depicted separately. Normalized log 10 expression values for each probe were compared between the normal and tumor samples using a two-tailed Wilcox matched-pairs sign rank test. The expression values for each data point were normalized to the average probe expression of the respective normal samples before plotting. Shaded boxes represent “normal” samples; open boxes represent “tumors.” (B) High Ubqln mRNA levels are associated with poor survival in lung adenocarcinoma. A set of 58 lung adenocarcinomas were separated into tertiles based on levels of Ubqln mRNA expression. Overall survival curves for the top and bottom tertiles were then plotted and compared using the Log-rank (Mantel-Cox) test. There is a significant association between the highest tertile of Ubqln RNA and poor prognosis (p = 0.0446). (C) IPA was performed on the 343 genes that were found to be significantly different between the Ubqln-high and Ubiqln-low human lung adenocarcinoma samples.
Fig. 7.
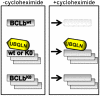
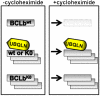
The regulation of concentrations of BCLb by Ubiquillin and ubiquitination. Treatment of cells expressing BCLbwt or BCLbK0 with cycloheximide lead to varied outcomes of the BCLb proteins depending on whether BCLb contains lysine residues or if Ubiquilin is present. Treatment of cells expressing BCLbwt with cycloheximide leads to rapid degradation of wild-type BCLb proteins (top). In contrast, treatment of cells expressing BCLbK0 with cycloheximide has no affect on BCLbK0 levels (bottom). When Ubqln is expressed with either BCLbwt or BCLbK0, there is a stabilization of BCLb proteins that is maintained following prolonged treatment with cycloheximide (middle).
Fig. P1.
The regulation of concentrations of BCLb by Ubiquillin and ubiquitination. Treatment of cells expressing BCLbwt or BCLbK0 with cycloheximide leads to varied outcomes of the BCLb proteins depending on whether BCLb contains lysine residues or if Ubiquilin is present. Treatment of cells expressing BCLbwt with cycloheximide leads to rapid degradation of BCLb proteins (top), whereas, there is no affect on BCLbK0 levels following treatment with cycloheximide (bottom). When Ubqln is expressed with either BCLbwt or BCLbK0, BCLb proteins remain stable during prolonged treatment with cycloheximide (middle).
Similar articles
- The STI and UBA Domains of UBQLN1 Are Critical Determinants of Substrate Interaction and Proteostasis.
Kurlawala Z, Shah PP, Shah C, Beverly LJ. Kurlawala Z, et al. J Cell Biochem. 2017 Aug;118(8):2261-2270. doi: 10.1002/jcb.25880. Epub 2017 Apr 25. J Cell Biochem. 2017. PMID: 28075048 Free PMC article. - MIR155 Regulation of Ubiquilin1 and Ubiquilin2: Implications in Cellular Protection and Tumorigenesis.
Yadav S, Singh N, Shah PP, Rowbotham DA, Malik D, Srivastav A, Shankar J, Lam WL, Lockwood WW, Beverly LJ. Yadav S, et al. Neoplasia. 2017 Apr;19(4):321-332. doi: 10.1016/j.neo.2017.02.001. Epub 2017 Mar 16. Neoplasia. 2017. PMID: 28315615 Free PMC article. - Decoding c-Myc networks of cell cycle and apoptosis regulated genes in a transgenic mouse model of papillary lung adenocarcinomas.
Ciribilli Y, Singh P, Spanel R, Inga A, Borlak J. Ciribilli Y, et al. Oncotarget. 2015 Oct 13;6(31):31569-92. doi: 10.18632/oncotarget.5035. Oncotarget. 2015. PMID: 26427040 Free PMC article. - Post-Transcriptional Regulation of Anti-Apoptotic BCL2 Family Members.
Cui J, Placzek WJ. Cui J, et al. Int J Mol Sci. 2018 Jan 20;19(1):308. doi: 10.3390/ijms19010308. Int J Mol Sci. 2018. PMID: 29361709 Free PMC article. Review. - Role of oxidative/nitrosative stress-mediated Bcl-2 regulation in apoptosis and malignant transformation.
Azad N, Iyer A, Vallyathan V, Wang L, Castranova V, Stehlik C, Rojanasakul Y. Azad N, et al. Ann N Y Acad Sci. 2010 Aug;1203:1-6. doi: 10.1111/j.1749-6632.2010.05608.x. Ann N Y Acad Sci. 2010. PMID: 20716276 Review.
Cited by
- PINK1 Content in Mitochondria is Regulated by ER-Associated Degradation.
Guardia-Laguarta C, Liu Y, Lauritzen KH, Erdjument-Bromage H, Martin B, Swayne TC, Jiang X, Przedborski S. Guardia-Laguarta C, et al. J Neurosci. 2019 Sep 4;39(36):7074-7085. doi: 10.1523/JNEUROSCI.1691-18.2019. Epub 2019 Jul 12. J Neurosci. 2019. PMID: 31300519 Free PMC article. - Dissecting the in vivo leukemogenic potency of BCLxl.
Saurabh K, Scherzer MT, Song A, Yip KW, Reed JC, Li C, Beverly LJ. Saurabh K, et al. J Leuk (Los Angel). 2014;2(5):158. doi: 10.4172/2329-6917.1000158. Epub 2014 Sep 24. J Leuk (Los Angel). 2014. PMID: 26636115 Free PMC article. - The novel ubiquitin ligase complex, SCF(Fbxw4), interacts with the COP9 signalosome in an F-box dependent manner, is mutated, lost and under-expressed in human cancers.
Lockwood WW, Chandel SK, Stewart GL, Erdjument-Bromage H, Beverly LJ. Lockwood WW, et al. PLoS One. 2013 May 2;8(5):e63610. doi: 10.1371/journal.pone.0063610. Print 2013. PLoS One. 2013. PMID: 23658844 Free PMC article. - HSP90-incorporating chaperome networks as biosensor for disease-related pathways in patient-specific midbrain dopamine neurons.
Kishinevsky S, Wang T, Rodina A, Chung SY, Xu C, Philip J, Taldone T, Joshi S, Alpaugh ML, Bolaender A, Gutbier S, Sandhu D, Fattahi F, Zimmer B, Shah SK, Chang E, Inda C, Koren J 3rd, Saurat NG, Leist M, Gross SS, Seshan VE, Klein C, Tomishima MJ, Erdjument-Bromage H, Neubert TA, Henrickson RC, Chiosis G, Studer L. Kishinevsky S, et al. Nat Commun. 2018 Oct 19;9(1):4345. doi: 10.1038/s41467-018-06486-6. Nat Commun. 2018. PMID: 30341316 Free PMC article. - Ushering in the cardiac role of Ubiquilin1.
Fang X, Trexler C, Chen J. Fang X, et al. J Clin Invest. 2018 Dec 3;128(12):5195-5197. doi: 10.1172/JCI124567. Epub 2018 Oct 22. J Clin Invest. 2018. PMID: 30352427 Free PMC article.
References
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
- Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. - PubMed
- Horvitz HR. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res. 1999;59(7 Suppl):1701s–1706s. - PubMed
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death . Nat Rev Mol Cell Biol. 2008;9:47–59. - PubMed
- Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 2P01CA094060-06A1/CA/NCI NIH HHS/United States
- P30 CA08748/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- P01 CA094060/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials