Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae - PubMed (original) (raw)
Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae
Simon R V Knott et al. Cell. 2012.
Abstract
The replication of eukaryotic chromosomes is organized temporally and spatially within the nucleus through epigenetic regulation of replication origin function. The characteristic initiation timing of specific origins is thought to reflect their chromatin environment or sub-nuclear positioning, however the mechanism remains obscure. Here we show that the yeast Forkhead transcription factors, Fkh1 and Fkh2, are global determinants of replication origin timing. Forkhead regulation of origin timing is independent of local levels or changes of transcription. Instead, we show that Fkh1 and Fkh2 are required for the clustering of early origins and their association with the key initiation factor Cdc45 in G1 phase, suggesting that Fkh1 and Fkh2 selectively recruit origins to emergent replication factories. Fkh1 and Fkh2 bind Fkh-activated origins, and interact physically with ORC, providing a plausible mechanism to cluster origins. These findings add a new dimension to our understanding of the epigenetic basis for differential origin regulation and its connection to chromosomal domain organization.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Figure 1
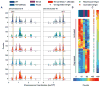
Analysis of early S-phase BrdU incorporation. A. BrdU incorporation plots of chromosomes III and VI are shown; plot colors and symbols correspond to the strain key above. Origins discussed in the text are boxed. B. Two-dimensional clustering of peak counts at Fkh-regulated origins is shown; columns (color-keyed above) correspond to strains and rows to origins. C. All detected origins (in rows) are arranged from maximum to minimum counts in WT, with the positions of Fkh-regulated origins indicated. See also Figure S1 and Data S1.
Figure 2
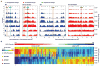
Temporal analysis of DNA replication by BrdU pulse-labeling. A. BrdU incorporation plots of chromosome III and a region of XV are shown. Origins discussed in the text are boxed. B. The matrix shows differences (WT-fkh1Δ fkh2ΔC) in BrdU incorporation (Δ M-value) at all Fkh-regulated origins (columns) across time (rows); the origins are arranged from left to right by their differences (WT-fkh1Δ fkh2ΔC) in BrdU incorporation in HU (Δ HU Counts). Specific origins are indicated below. See also Figure S2 and Data S2.
Figure 3
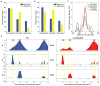
Analysis of Fkh1 and Fkh2 binding sites near origins. A and B. Frequencies of expected and actual Fkh1 (A) and Fkh2 (B) consensus binding sites near Fkh-activated, Fkh-unregulated, and Fkh-repressed origins are shown. C. Frequency distribution plots of Fkh1 and Fkh2 consensus binding sites relative to ACS position are shown. D. M-values for BrdU-IP-chip and for ChIP-chip of Fkh1 and ORC binding along the ARS305 region in WT cells harboring ARS305 or ars305Δ2BS.
Figure 4
Transcription analysis surrounding Fkh-regulated origins in unsynchronized and G1-synchronized cells. RNA-Seq (A) and Rpb3 ChIP-Seq (B) read counts of WT, fkh1Δ fkh2ΔC, and WT-fkh1Δ fkh2ΔC differences (Δ), within 10kb of each Fkh-regulated origin, are aligned by each origin’s predicted or verified ACS. Origins are grouped according to the orientation of the flanking genes, and arranged by differences (WT-fkh1Δ fkh2ΔC) in BrdU incorporation in HU (Δ HU Counts).
Figure 5
Genome-wide binding of replication initiation factors to Fkh-regulated origins. A. M-values from ChIP-chip analysis of ORC, Mcm2+4, and Cdc45 at Fkh-regulated origins (in rows) are arranged by differences (WT-fkh1Δ fkh2ΔC) in BrdU incorporation in HU (Δ HU Counts). B. Venn diagram of Cdc45 binding within different origin classes is shown.
Figure 6
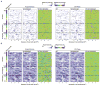
Chromosome-conformation capture analyses of origin interactions. A. Two-dimensional clustering of origin-origin interaction frequencies is shown, with origins in columns and rows of the matrix. Columns to the right indicate Cdc45 ChIP-chip binding, average BrdU ΔHU-counts, and ΔBrdU-pulse M-values. The top 5% (based on p values) of Fkh-activated and Fkh-repressed origins are indicated. B. Venn diagram of overlap between experimental replicates is shown. C. Plots of the ARS607 region including relevant XbaI sites are shown. See also Figure S4.
Figure 7
Co-IP of Fkh1 with ORC. Soluble extracts from FKH1-MYC (lanes 1, 3, 5–8) and untagged (lanes 2, 4) cells were subjected to IP with anti-Myc antibody (A) and anti-ORC antibody (B and C). IPs were analyzed by immunoblotting with anti-Myc and anti-ORC antibodies. C. Ethidium bromide (EtBr) was included in the IPs at 10, 40, and 100 μg/mL in lanes 6, 7, and 8, respectively. ORC protein was included as standard.
Similar articles
- The budding yeast Fkh1 Forkhead associated (FHA) domain promotes a G1-chromatin state and the activity of chromosomal DNA replication origins.
Hoggard T, Chacin E, Hollatz AJ, Kurat CF, Fox CA. Hoggard T, et al. PLoS Genet. 2024 Aug 5;20(8):e1011366. doi: 10.1371/journal.pgen.1011366. eCollection 2024 Aug. PLoS Genet. 2024. PMID: 39102423 Free PMC article. - Conserved forkhead dimerization motif controls DNA replication timing and spatial organization of chromosomes in S. cerevisiae.
Ostrow AZ, Kalhor R, Gan Y, Villwock SK, Linke C, Barberis M, Chen L, Aparicio OM. Ostrow AZ, et al. Proc Natl Acad Sci U S A. 2017 Mar 21;114(12):E2411-E2419. doi: 10.1073/pnas.1612422114. Epub 2017 Mar 6. Proc Natl Acad Sci U S A. 2017. PMID: 28265091 Free PMC article. - Quantitative BrdU immunoprecipitation method demonstrates that Fkh1 and Fkh2 are rate-limiting activators of replication origins that reprogram replication timing in G1 phase.
Peace JM, Villwock SK, Zeytounian JL, Gan Y, Aparicio OM. Peace JM, et al. Genome Res. 2016 Mar;26(3):365-75. doi: 10.1101/gr.196857.115. Epub 2016 Jan 4. Genome Res. 2016. PMID: 26728715 Free PMC article. - Behavior of replication origins in Eukaryota - spatio-temporal dynamics of licensing and firing.
Musiałek MW, Rybaczek D. Musiałek MW, et al. Cell Cycle. 2015;14(14):2251-64. doi: 10.1080/15384101.2015.1056421. Epub 2015 Jun 1. Cell Cycle. 2015. PMID: 26030591 Free PMC article. Review. - Chromatin proteins involved in the initiation of DNA replication.
Rowles A, Blow JJ. Rowles A, et al. Curr Opin Genet Dev. 1997 Apr;7(2):152-7. doi: 10.1016/s0959-437x(97)80123-2. Curr Opin Genet Dev. 1997. PMID: 9115430 Review.
Cited by
- The budding yeast Fkh1 Forkhead associated (FHA) domain promotes a G1-chromatin state and the activity of chromosomal DNA replication origins.
Hoggard T, Chacin E, Hollatz AJ, Kurat CF, Fox CA. Hoggard T, et al. PLoS Genet. 2024 Aug 5;20(8):e1011366. doi: 10.1371/journal.pgen.1011366. eCollection 2024 Aug. PLoS Genet. 2024. PMID: 39102423 Free PMC article. - Sustained inactivation of the Polycomb PRC1 complex induces DNA repair defects and genomic instability in epigenetic tumors.
Rawal CC, Loubiere V, Butova NL, Gracia J, Parreno V, Merigliano C, Martinez AM, Cavalli G, Chiolo I. Rawal CC, et al. Histochem Cell Biol. 2024 Jul;162(1-2):133-147. doi: 10.1007/s00418-024-02302-z. Epub 2024 Jun 18. Histochem Cell Biol. 2024. PMID: 38888809 Free PMC article. - Sustained inactivation of the Polycomb PRC1 complex induces DNA repair defects and genomic instability in epigenetic tumors.
Rawal CC, Loubiere V, Butova NL, Garcia J, Parreno V, Martinez AM, Cavalli G, Chiolo I. Rawal CC, et al. Res Sq [Preprint]. 2024 Apr 24:rs.3.rs-4289524. doi: 10.21203/rs.3.rs-4289524/v1. Res Sq. 2024. PMID: 38746379 Free PMC article. Updated. Preprint. - The budding yeast Fkh1 Forkhead associated (FHA) domain promoted a G1-chromatin state and the activity of chromosomal DNA replication origins.
Hoggard T, Chacin E, Hollatz AJ, Kurat CF, Fox CA. Hoggard T, et al. bioRxiv [Preprint]. 2024 Jun 7:2024.02.16.580712. doi: 10.1101/2024.02.16.580712. bioRxiv. 2024. PMID: 38405780 Free PMC article. Updated. Preprint. - The origin recognition complex requires chromatin tethering by a hypervariable intrinsically disordered region that is functionally conserved from sponge to man.
Adiji OA, McConnell BS, Parker MW. Adiji OA, et al. Nucleic Acids Res. 2024 May 8;52(8):4344-4360. doi: 10.1093/nar/gkae122. Nucleic Acids Res. 2024. PMID: 38381902 Free PMC article.
References
- Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. - PubMed
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM065494-09/GM/NIGMS NIH HHS/United States
- R01 GM065494/GM/NIGMS NIH HHS/United States
- 3R01-GM065494-S1/GM/NIGMS NIH HHS/United States
- 5R01-GM065494/GM/NIGMS NIH HHS/United States
- P50-HG02790/HG/NHGRI NIH HHS/United States
- P50 HG002790/HG/NHGRI NIH HHS/United States
- R01 GM065494-07S1/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials