Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities - PubMed (original) (raw)
. 2012 Jan 22;482(7383):98-102.
doi: 10.1038/nature10814.
Murim Choi, Keith A Choate, Carol J Nelson-Williams, Anita Farhi, Hakan R Toka, Irina R Tikhonova, Robert Bjornson, Shrikant M Mane, Giacomo Colussi, Marcel Lebel, Richard D Gordon, Ben A Semmekrot, Alain Poujol, Matti J Välimäki, Maria E De Ferrari, Sami A Sanjad, Michael Gutkin, Fiona E Karet, Joseph R Tucci, Jim R Stockigt, Kim M Keppler-Noreuil, Craig C Porter, Sudhir K Anand, Margo L Whiteford, Ira D Davis, Stephanie B Dewar, Alberto Bettinelli, Jeffrey J Fadrowski, Craig W Belsha, Tracy E Hunley, Raoul D Nelson, Howard Trachtman, Trevor R P Cole, Maury Pinsk, Detlef Bockenhauer, Mohan Shenoy, Priya Vaidyanathan, John W Foreman, Majid Rasoulpour, Farook Thameem, Hania Z Al-Shahrouri, Jai Radhakrishnan, Ali G Gharavi, Beatrice Goilav, Richard P Lifton
Affiliations
- PMID: 22266938
- PMCID: PMC3278668
- DOI: 10.1038/nature10814
Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities
Lynn M Boyden et al. Nature. 2012.
Abstract
Hypertension affects one billion people and is a principal reversible risk factor for cardiovascular disease. Pseudohypoaldosteronism type II (PHAII), a rare Mendelian syndrome featuring hypertension, hyperkalaemia and metabolic acidosis, has revealed previously unrecognized physiology orchestrating the balance between renal salt reabsorption and K(+) and H(+) excretion. Here we used exome sequencing to identify mutations in kelch-like 3 (KLHL3) or cullin 3 (CUL3) in PHAII patients from 41 unrelated families. KLHL3 mutations are either recessive or dominant, whereas CUL3 mutations are dominant and predominantly de novo. CUL3 and BTB-domain-containing kelch proteins such as KLHL3 are components of cullin-RING E3 ligase complexes that ubiquitinate substrates bound to kelch propeller domains. Dominant KLHL3 mutations are clustered in short segments within the kelch propeller and BTB domains implicated in substrate and cullin binding, respectively. Diverse CUL3 mutations all result in skipping of exon 9, producing an in-frame deletion. Because dominant KLHL3 and CUL3 mutations both phenocopy recessive loss-of-function KLHL3 mutations, they may abrogate ubiquitination of KLHL3 substrates. Disease features are reversed by thiazide diuretics, which inhibit the Na-Cl cotransporter in the distal nephron of the kidney; KLHL3 and CUL3 are expressed in this location, suggesting a mechanistic link between KLHL3 and CUL3 mutations, increased Na-Cl reabsorption, and disease pathogenesis. These findings demonstrate the utility of exome sequencing in disease gene identification despite the combined complexities of locus heterogeneity, mixed models of transmission and frequent de novo mutation, and establish a fundamental role for KLHL3 and CUL3 in blood pressure, K(+) and pH homeostasis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
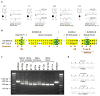
Figure 1. Recessive and dominant KLHL3 mutations in PHAII kindreds
a–b, Representative kindreds demonstrating recessive (a) and dominant (b) KLHL3 mutations (all 24 kindreds are shown in Supplementary Figs. 3–4). Affected, unaffected, and phenotype-undetermined subjects are denoted by black, white, and gray symbols, respectively. KLHL3 alleles are denoted by ‘+’ (wild-type), ‘d’ (recessive mutation), and ‘D’ (dominant mutation). Sequence traces show wild-type (wt) and mutant (*) alleles and encoded amino acids. c, KLHL3 protein sequence. Colored bars indicate BTB domain (lavender), BACK domain (peach), and Kelch propeller blades (B1-B6, gray) with β-strands ‘a’–‘d’ in yellow, red, green, and blue respectively. Recessive (aqua) and dominant (pink) mutations are shown; recurrences indicated by numbers. d, Kelch propeller schematic, from KLHL2 crystal structure. β-strands colored as in c; dominant mutations indicated. e, CRL schematic, comprising a BTB-Kelch protein (KLHL3), CUL3, and a ubiquitin transfer-mediating RING protein, with substrate bound via the Kelch propeller. Complex shown as a dimer.
Figure 2. Dominant CUL3 mutations in PHAII kindreds cause skipping of exon 9
a, Representative kindreds demonstrating CUL3 mutations, depicted as in Fig. 1 (all 17 kindreds are shown in Supplementary Fig. 6). b, CUL3 mutation locations. Consensus splicing sequences, and corresponding wildtype CUL3 sequences within intron 8, exon 9, and intron 9 are shown; invariant bases (green) and consensus homology (yellow) are indicated. Positions numbered relative to splice sites and first base of the exonic splice (ES) enhancer. Mutations shown in red; recurrences indicated by numbers. c, RT-PCR of spliced RNA. Wild-type CUL3 constructs produce a single product including exons 8, 9, and 10 (844 bp); all nine mutants tested produce a predominant product that skips exon 9 (673 bp). d, Representative RT-PCR sequences. Wild-type construct produces cDNA with properly spliced junctions between exons 8–9 (top) and 9–10 (middle), while mutant construct [splice donor g(+1)c] produces cDNA joining exon 8 to exon 10 (bottom).
Figure 3. KLHL3 expression in the kidney
Mouse kidney sections stained with antibodies to KLHL3 (red), TRPM6 (a marker of the DCT, green) and AQP2 (a marker of the CD, blue). Scale bars 25 μm. a–c, Staining for KLHL3 (a), TRPM6 (b), and the merged image (c) demonstrates KLHL3 expression in the DCT with apical localization (arrowheads). d–f, Staining for KLHL3 (d), AQP2 (e), and the merged image (f) demonstrates KLHL3 expression in CD.
Comment in
- Exome sequencing to identify novel genes in hypertension.
Musunuru K. Musunuru K. Circ Cardiovasc Genet. 2012 Apr 1;5(2):267-8. doi: 10.1161/CIRCGENETICS.112.963256. Circ Cardiovasc Genet. 2012. PMID: 22511709 No abstract available.
Similar articles
- Detection of mutations in KLHL3 and CUL3 in families with FHHt (familial hyperkalaemic hypertension or Gordon's syndrome).
Glover M, Ware JS, Henry A, Wolley M, Walsh R, Wain LV, Xu S, Van't Hoff WG, Tobin MD, Hall IP, Cook S, Gordon RD, Stowasser M, O'Shaughnessy KM. Glover M, et al. Clin Sci (Lond). 2014 May;126(10):721-6. doi: 10.1042/CS20130326. Clin Sci (Lond). 2014. PMID: 24266877 Free PMC article. - The CUL3-KLHL3 E3 ligase complex mutated in Gordon's hypertension syndrome interacts with and ubiquitylates WNK isoforms: disease-causing mutations in KLHL3 and WNK4 disrupt interaction.
Ohta A, Schumacher FR, Mehellou Y, Johnson C, Knebel A, Macartney TJ, Wood NT, Alessi DR, Kurz T. Ohta A, et al. Biochem J. 2013 Apr 1;451(1):111-22. doi: 10.1042/BJ20121903. Biochem J. 2013. PMID: 23387299 Free PMC article. - Kelch-like 3 and Cullin 3 regulate electrolyte homeostasis via ubiquitination and degradation of WNK4.
Shibata S, Zhang J, Puthumana J, Stone KL, Lifton RP. Shibata S, et al. Proc Natl Acad Sci U S A. 2013 May 7;110(19):7838-43. doi: 10.1073/pnas.1304592110. Epub 2013 Apr 1. Proc Natl Acad Sci U S A. 2013. PMID: 23576762 Free PMC article. - Kelch-like protein 3 in human disease and therapy.
Lin Y, Li Q, Jin X. Lin Y, et al. Mol Biol Rep. 2022 Oct;49(10):9813-9824. doi: 10.1007/s11033-022-07487-x. Epub 2022 May 18. Mol Biol Rep. 2022. PMID: 35585379 Review. - Kelch-like 3/Cullin 3 ubiquitin ligase complex and WNK signaling in salt-sensitive hypertension and electrolyte disorder.
Sohara E, Uchida S. Sohara E, et al. Nephrol Dial Transplant. 2016 Sep;31(9):1417-24. doi: 10.1093/ndt/gfv259. Epub 2015 Jul 6. Nephrol Dial Transplant. 2016. PMID: 26152401 Review.
Cited by
- Clinical and laboratory approaches in the diagnosis of renal tubular acidosis.
Santos F, Ordóñez FA, Claramunt-Taberner D, Gil-Peña H. Santos F, et al. Pediatr Nephrol. 2015 Dec;30(12):2099-107. doi: 10.1007/s00467-015-3083-9. Epub 2015 Apr 1. Pediatr Nephrol. 2015. PMID: 25823989 Review. - Application of whole exome sequencing to identify disease-causing variants in inherited human diseases.
Goh G, Choi M. Goh G, et al. Genomics Inform. 2012 Dec;10(4):214-9. doi: 10.5808/GI.2012.10.4.214. Epub 2012 Dec 31. Genomics Inform. 2012. PMID: 23346032 Free PMC article. - Panel sequencing distinguishes monogenic forms of nephritis from nephrosis in children.
Schapiro D, Daga A, Lawson JA, Majmundar AJ, Lovric S, Tan W, Warejko JK, Fessi I, Rao J, Airik M, Gee HY, Schneider R, Widmeier E, Hermle T, Ashraf S, Jobst-Schwan T, van der Ven AT, Nakayama M, Shril S, Braun DA, Hildebrandt F. Schapiro D, et al. Nephrol Dial Transplant. 2019 Mar 1;34(3):474-485. doi: 10.1093/ndt/gfy050. Nephrol Dial Transplant. 2019. PMID: 30295827 Free PMC article. - Inwardly rectifying K+ channels 4.1 and 5.1 (Kir4.1/Kir5.1) in the renal distal nephron.
Wang WH, Lin DH. Wang WH, et al. Am J Physiol Cell Physiol. 2022 Aug 1;323(2):C277-C288. doi: 10.1152/ajpcell.00096.2022. Epub 2022 Jun 27. Am J Physiol Cell Physiol. 2022. PMID: 35759440 Free PMC article. Review. - Protein phosphatase 1 inhibitor-1 deficiency reduces phosphorylation of renal NaCl cotransporter and causes arterial hypotension.
Picard N, Trompf K, Yang CL, Miller RL, Carrel M, Loffing-Cueni D, Fenton RA, Ellison DH, Loffing J. Picard N, et al. J Am Soc Nephrol. 2014 Mar;25(3):511-22. doi: 10.1681/ASN.2012121202. Epub 2013 Nov 14. J Am Soc Nephrol. 2014. PMID: 24231659 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK079310/DK/NIDDK NIH HHS/United States
- P30-DK079310/DK/NIDDK NIH HHS/United States
- KL2 RR024138/RR/NCRR NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- UL1-RR024139/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases