Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event - PubMed (original) (raw)
. 2012 Jan 29;44(3):352-5.
doi: 10.1038/ng.1072.
Angela B Brueggemann, Teresa Street, Robert E Gertz Jr, Chris C A Spencer, Thien Ho, Eleni Giannoulatou, Ruth Link-Gelles, Rosalind M Harding, Bernard Beall, Tim E A Peto, Matthew R Moore, Peter Donnelly, Derrick W Crook, Rory Bowden
Affiliations
- PMID: 22286217
- PMCID: PMC3303117
- DOI: 10.1038/ng.1072
Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event
Tanya Golubchik et al. Nat Genet. 2012.
Abstract
Streptococcus pneumoniae ('pneumococcus') causes an estimated 14.5 million cases of serious disease and 826,000 deaths annually in children under 5 years of age(1). The highly effective introduction of the PCV7 pneumococcal vaccine in 2000 in the United States(2,3) provided an unprecedented opportunity to investigate the response of an important pathogen to widespread, vaccine-induced selective pressure. Here, we use array-based sequencing of 62 isolates from a US national monitoring program to study five independent instances of vaccine escape recombination(4), showing the simultaneous transfer of multiple and often large (up to at least 44 kb) DNA fragments. We show that one such new strain quickly became established, spreading from east to west across the United States. These observations clarify the roles of recombination and selection in the population genomics of pneumococcus and provide proof of principle of the considerable value of combining genomic and epidemiological information in the surveillance and enhanced understanding of infectious diseases.
Figures
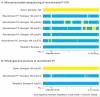
Figure 1
Resequencing of pneumococcal vaccine escape recombinants: comparison of recombinant and putative recipient and donor sequences. (A) Vaccine escape recombinants P1-P5 were resequenced using the Affymetrix CustomSeq platform, a microarray-based approach covering a selected 12% of the pneumococcus genome (300 kbp). The genomes are coloured by inferred origin of genomic intervals (blue - recipient; yellow - donor). The capsular locus transferred in a different form in each recombinant is evident as a large yellow block centred on the same location towards the left of each genome. Other yellow blocks indicate extra transferred sequences. The recipient genomes in all cases were highly similar. (B) Whole-genome structure of recombinant P1. Representatives of recombinant P1 and its best-matching prospective parents were sequenced on the Illumina GAIIx platform. Sequence reads were assembled de novo and aligned to the TIGR4 reference sequence and a heuristic algorithm was implemented in which three or more donor-like single nucleotide variants encompassing no more than one recipient-like variant was used to identify donor genomic fragments (blue - recipient; yellow - donor). This approach detected 15 imports of ≥50bp, including 8 that did not overlap previously detected fragments. The scale in (B) is different to that in (A) because the reference sequences differ slightly. In each panel the smallest fragments (triangles) have been magnified to a minimum size of 5kb for visibility.
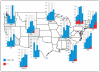
Figure 2
Spread of P1 vaccine escape recombinant through space and time. Incidence of P1 vaccine escape recombinants (red) and other 19A (blue) as a proportion of all pneumococci in invasive pneumococcal disease among children under 5 years of age. Data are shown for the years 2003-2007; left to right panels for each state in 10 ABCs monitoring states. Data from 2003 in New Mexico are missing (grey box) because surveillance started there in 2004. P1 was first detected in New York and Connecticut from where it has spread westwards.
Similar articles
- Vaccine escape recombinants emerge after pneumococcal vaccination in the United States.
Brueggemann AB, Pai R, Crook DW, Beall B. Brueggemann AB, et al. PLoS Pathog. 2007 Nov;3(11):e168. doi: 10.1371/journal.ppat.0030168. PLoS Pathog. 2007. PMID: 18020702 Free PMC article. - Prediction of post-PCV13 pneumococcal evolution using invasive disease data enhanced by inverse-invasiveness weighting.
Qiu X, McGee L, Hammitt L, Grant LR, O'Brien KL, Hanage WP, Lipsitch M. Qiu X, et al. mBio. 2024 Oct 16;15(10):e0335523. doi: 10.1128/mbio.03355-23. Epub 2024 Aug 29. mBio. 2024. PMID: 39207103 Free PMC article. - Phylogeography and resistome of pneumococcal meningitis in West Africa before and after vaccine introduction.
Senghore M, Tientcheu PE, Worwui AK, Jarju S, Okoi C, Suso SMS, Foster-Nyarko E, Ebruke C, Sonko M, Kourna MH, Agossou J, Tsolenyanu E, Renner LA, Ansong D, Sanneh B, Cisse CB, Boula A, Miwanda B, Lo SW, Gladstone RA, Schwartz S, Hawkins P, McGee L, Klugman KP, Breiman RF, Bentley SD, Mwenda JM, Kwambana-Adams BA, Antonio M. Senghore M, et al. Microb Genom. 2021 Jul;7(7):000506. doi: 10.1099/mgen.0.000506. Microb Genom. 2021. PMID: 34328412 Free PMC article. - Global genomic pathogen surveillance to inform vaccine strategies: a decade-long expedition in pneumococcal genomics.
Bentley SD, Lo SW. Bentley SD, et al. Genome Med. 2021 May 17;13(1):84. doi: 10.1186/s13073-021-00901-2. Genome Med. 2021. PMID: 34001237 Free PMC article. Review. - Natural transformation and genome evolution in Streptococcus pneumoniae.
Straume D, Stamsås GA, Håvarstein LS. Straume D, et al. Infect Genet Evol. 2015 Jul;33:371-80. doi: 10.1016/j.meegid.2014.10.020. Epub 2014 Nov 4. Infect Genet Evol. 2015. PMID: 25445643 Review.
Cited by
- Genomic analyses of pneumococci reveal a wide diversity of bacteriocins - including pneumocyclicin, a novel circular bacteriocin.
Bogaardt C, van Tonder AJ, Brueggemann AB. Bogaardt C, et al. BMC Genomics. 2015 Jul 28;16(1):554. doi: 10.1186/s12864-015-1729-4. BMC Genomics. 2015. PMID: 26215050 Free PMC article. - Development of a Dominant Negative Competence-Stimulating Peptide (dnCSP) that Attenuates Streptococcus pneumoniae Infectivity in a Mouse Model of Acute Pneumonia.
Koirala B, Lin J, Lau GW, Tal-Gan Y. Koirala B, et al. Chembiochem. 2018 Nov 16;19(22):2380-2386. doi: 10.1002/cbic.201800505. Epub 2018 Oct 23. Chembiochem. 2018. PMID: 30211457 Free PMC article. - Differences in genetic flux in invasive Streptococcus pneumoniae associated with bacteraemia and meningitis.
Mutua TM, Kulohoma BW. Mutua TM, et al. Heliyon. 2022 Dec 15;8(12):e12229. doi: 10.1016/j.heliyon.2022.e12229. eCollection 2022 Dec. Heliyon. 2022. PMID: 36593853 Free PMC article. - Pandemic-scale phylogenomics reveals the SARS-CoV-2 recombination landscape.
Turakhia Y, Thornlow B, Hinrichs A, McBroome J, Ayala N, Ye C, Smith K, De Maio N, Haussler D, Lanfear R, Corbett-Detig R. Turakhia Y, et al. Nature. 2022 Sep;609(7929):994-997. doi: 10.1038/s41586-022-05189-9. Epub 2022 Aug 11. Nature. 2022. PMID: 35952714 Free PMC article. - Insights from genomics into bacterial pathogen populations.
Wilson DJ. Wilson DJ. PLoS Pathog. 2012 Sep;8(9):e1002874. doi: 10.1371/journal.ppat.1002874. Epub 2012 Sep 6. PLoS Pathog. 2012. PMID: 22969423 Free PMC article. Review.
References
- O’Brien KL, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374:893–902. - PubMed
- Whitney CG, et al. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N Engl J Med. 2003;348:1737–46. - PubMed
- Pilishvili T, et al. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis. 2010;201:32–41. - PubMed
- Eskola J, et al. A randomized, prospective field trial of a conjugate vaccine in the protection of infants and young children against invasive Haemophilus influenzae type b disease. N Engl J Med. 1990;323:1381–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 083511/Z/07/Z/WT_/Wellcome Trust/United Kingdom
- 079126/WT_/Wellcome Trust/United Kingdom
- G0800778/MRC_/Medical Research Council/United Kingdom
- 083511/WT_/Wellcome Trust/United Kingdom
- G0800778(88305)/MRC_/Medical Research Council/United Kingdom
- 079126/Z/06/Z/WT_/Wellcome Trust/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- 090532/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- 087646/WT_/Wellcome Trust/United Kingdom
- 087646/2/08/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical