Cancer stem cells: impact, heterogeneity, and uncertainty - PubMed (original) (raw)
Review
Cancer stem cells: impact, heterogeneity, and uncertainty
Jeffrey A Magee et al. Cancer Cell. 2012.
Abstract
The differentiation of tumorigenic cancer stem cells into nontumorigenic cancer cells confers heterogeneity to some cancers beyond that explained by clonal evolution or environmental differences. In such cancers, functional differences between tumorigenic and nontumorigenic cells influence response to therapy and prognosis. However, it remains uncertain whether the model applies to many, or few, cancers due to questions about the robustness of cancer stem cell markers and the extent to which existing assays underestimate the frequency of tumorigenic cells. In cancers with rapid genetic change, reversible changes in cell states, or biological variability among patients, the stem cell model may not be readily testable.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Figure 1. Sources of heterogeneity within cancer
A) Heterogeneity can arise within tumors through stochastic genetic (Nowell, 1976) and epigenetic (Baylin and Jones, 2011) changes that confer heritable phenotypic and functional differences upon cancer cells. This process is known as clonal evolution because the genetic/epigenetic changes are subject to selection within tumors. This process tends to lead to more aggressive cancers over time; however, some cancer cells (grey) would be predicted to lose their tumorigenic capacity as a consequence of disadvantageous genetic changes. B) Heterogeneity can arise in response to extrinsic environmental differences within tumors: cancer cells (blue) adjacent to blood vessels (red) are different from cancer cells further from blood vessels (white) (Charles et al., 2010). The differences are shown as being reversible, though environmental differences could also cause irreversible changes in cancer cell properties. C) Cancers that follow the stem cell model contain intrinsically different subpopulations of tumorigenic (red) and non-tumorigenic cells (yellow and green) organized in a hierarchy in which a minority population of tumorigenic cells gives rise to phenotypically diverse non-tumorigenic cells. Non-tumorigenic cells are thought to compose the bulk of tumors but have little capacity to contribute to cancer progression (Dick, 2008; Reya et al., 2001; Shackleton et al., 2009). Tumorigenic cells can be serially transplanted, re-establishing phenotypic heterogeneity with each passage. D) Cancers that follow the stem cell model are also subject to clonal evolution as well as heterogeneity from environmental differences within tumors. Thus, these sources of heterogeneity are not mutually exclusive and may each apply to variable extents depending on the cancer.
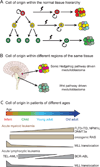
Figure 2. Sources of heterogeneity among cancers
Differences in the cell-of-origin can directly and indirectly influence the phenotype of tumorigenic cells and, perhaps, whether or not the cancer is hierarchically organized. A) Different cell types in a stem/progenitor cell hierarchy within a normal tissue may be transformed into cancer cells. The properties of the cell-of-origin influence the types of mutations that are competent to transform and the properties of the resulting cancer (Huntly et al., 2004; Wang et al., 2010). (B) Spatial differences in the identity of the cell-of-origin within tissues influence the types of mutations that are competent to transform and the properties of the resulting cancer (Gibson et al., 2010; Johnson et al., 2010). (C) Temporal differences in the cell-of-origin also influence the types of mutations that are competent to transform and the properties of the resulting cancer (Magee and Morrison, unpublished data), consistent with the observation that the driver mutation spectrum changes with age in patients (Downing and Shannon, 2002)(see text for references regarding age-related changes in the incidence of specific mutations).
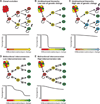
Figure 3. Variation among leukemias in the degree and nature of hierarchical organization
Although most AMLs follow a cancer stem cell model, the surface marker phenotypes of the leukemogenic cells vary from patient to patient. (A–C) Different oncogenic mutations can transform cells at different levels within the hematopoiesis hierarchy (Wang et al., 2010), potentially influencing the frequency, spectrum, and phenotype of cells with leukemogenic potential. Interconversion between leukemogenic cell populations would allow any population to recapitulate the heterogeneity of the leukemogenic cell pool (Eppert et al., 2011; Sarry et al., 2011). (D) Some mutations, such as MLL-AF9 translocations, can confer leukemogenic activity upon restricted progenitors (Cozzio et al., 2003; Huntly et al., 2004; Krivtsov et al., 2006; Zhao et al., 2010). (E) If multiple populations have leukemogenic capacity but do not interconvert, then only the most immature cells can recapitulate the full heterogeneity of the parent leukemia but multiple levels of the hierarchy may be able to drive disease progression (Goardon et al., 2011). (F) In some ALLs many cells have leukemogenic activity despite heterogeneity in marker expression (le Viseur et al., 2008; Williams et al., 2007).
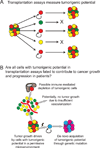
Figure 4. Fate versus potential in cancer
A) Potential describes what cells can do in a permissive environment. The cancer stem cell model, and the transplantation assays (black arrows) on which it is largely based, address the potential of cancer cells to form tumors. B) Fate reflects what cells actually do in a specific environment. In the context of cancer, the question is which cells are fated to contribute to tumor growth and disease progression in their actual environment in the patient. Many of the cells that have the potential to form tumors upon transplantation may not be fated to contribute to disease progression in a particular patient because they are not in a permissive environment. For example, some cancer cells undergo cell death due to hypoxia or immune effector activity. Some of these cells might have the potential to form a tumor if transplanted into another environment, but are fated to undergo cell death in the tumor environment in which they actually reside in the patient. There may also be cells that lack the ability to form a tumor upon transplantation but that are nonetheless fated to contribute to disease progression in the patient, such as if they acquire a new mutation that increases their proliferation. Transplantation assays only assess potential, not fate in a patient, and therefore should attempt to detect the full range of cells with the potential to form tumors. No transplantation assay mimics the environment within patient tissues and such assays are neither designed nor capable of assessing cell fate in patients. Consequently, very little is known about the spectrum of cells fated to contribute to tumor growth and disease progression in patients, or the extent to which it overlaps with the spectrum of cells that can form a tumor upon transplantation.
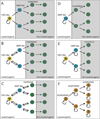
Figure 5. The influences of genetic change and reversible transitions in cell states on hierarchical organization in cancer
A) According to the clonal evolution model many cancer cells have tumorigenic potential (circular self-renewal arrows) and heterogeneity arises through stochastic genetic/epigenetic changes (lightning bolt). Changes in cell phenotype are not necessarily associated with changes in tumorigenic potential. B) For cancers that follow a stem cell model in which only the cells at the top of the hierarchy retain tumorigenic capacity, the differentiation of these cells into non-tumorigenic progeny creates tumor heterogeneity. Differentiation is associated with a loss of tumorigenic potential. C) For cancers with a high rate of genetic change, clones of cells within the hierarchy depicted in panel B may acquire tumorigenic potential as a consequence of new mutations. Phenotypic changes are sometimes associated with changes in tumorigenic potential and sometimes not. Note that it would be difficult to experimentally distinguish this model from the model in panel A. D) A cancer that is hierarchically organized according to the cancer stem cell model but in which non-tumorigenic cells can inefficiently revert to higher levels of the hierarchy. In this case, tumorigenic cells could be enriched or depleted using markers but “non-tumorigenic” cells from the bottom of the hierarchy would always retain some tumorigenic capacity due to their ability to revert to tumorigenic states. E) A hierarchically organized cancer in which cells readily and reversibly interconvert between tumorigenic (red) and non-tumorigenic (yellow and green) states. Note that it would be difficult to experimentally distinguish case (E) from case (C). In cancers that are genetically unstable or subject to efficient reversible cell transitions, the cancer stem cell model may be untestable as it may be difficult to experimentally distinguish from cancers in which there is no hierarchy but where heterogeneity arises through clonal evolution.
Similar articles
- Tumour heterogeneity and cancer cell plasticity.
Meacham CE, Morrison SJ. Meacham CE, et al. Nature. 2013 Sep 19;501(7467):328-37. doi: 10.1038/nature12624. Nature. 2013. PMID: 24048065 Free PMC article. Review. - Evolution of cancer stem cells.
Bapat SA. Bapat SA. Semin Cancer Biol. 2007 Jun;17(3):204-13. doi: 10.1016/j.semcancer.2006.05.001. Epub 2006 May 20. Semin Cancer Biol. 2007. PMID: 16787749 Review. - Cancer stem cell marker phenotypes are reversible and functionally homogeneous in a preclinical model of pancreatic cancer.
Dosch JS, Ziemke EK, Shettigar A, Rehemtulla A, Sebolt-Leopold JS. Dosch JS, et al. Cancer Res. 2015 Nov 1;75(21):4582-92. doi: 10.1158/0008-5472.CAN-14-2793. Epub 2015 Sep 10. Cancer Res. 2015. PMID: 26359451 Free PMC article. - Roots and stems: stem cells in cancer.
Polyak K, Hahn WC. Polyak K, et al. Nat Med. 2006 Mar;12(3):296-300. doi: 10.1038/nm1379. Nat Med. 2006. PMID: 16520777 - Recurrence cancer stem cells--made by cell fusion?
Dittmar T, Nagler C, Schwitalla S, Reith G, Niggemann B, Zänker KS. Dittmar T, et al. Med Hypotheses. 2009 Oct;73(4):542-7. doi: 10.1016/j.mehy.2009.05.044. Epub 2009 Jun 28. Med Hypotheses. 2009. PMID: 19564079
Cited by
- Differential characteristics of CD133(+) and CD133 (-) Jurkat cells.
Anbarlou A, Atashi A, Soleimani M, AkhavanRahnama M, Bohloli M, Mossahebi-Mohammadi M. Anbarlou A, et al. In Vitro Cell Dev Biol Anim. 2015 Jun;51(6):556-61. doi: 10.1007/s11626-015-9869-z. Epub 2015 Jan 29. In Vitro Cell Dev Biol Anim. 2015. PMID: 25630537 - Targeting ALDH1A1 by disulfiram/copper complex inhibits non-small cell lung cancer recurrence driven by ALDH-positive cancer stem cells.
Liu X, Wang L, Cui W, Yuan X, Lin L, Cao Q, Wang N, Li Y, Guo W, Zhang X, Wu C, Yang J. Liu X, et al. Oncotarget. 2016 Sep 6;7(36):58516-58530. doi: 10.18632/oncotarget.11305. Oncotarget. 2016. PMID: 27542268 Free PMC article. - Does Cancer Biology Rely on Parrondo's Principles?
Capp JP, Nedelcu AM, Dujon AM, Roche B, Catania F, Ujvari B, Alix-Panabières C, Thomas F. Capp JP, et al. Cancers (Basel). 2021 May 3;13(9):2197. doi: 10.3390/cancers13092197. Cancers (Basel). 2021. PMID: 34063648 Free PMC article. Review. - Biology of lung cancer: genetic mutation, epithelial-mesenchymal transition, and cancer stem cells.
Aoi T. Aoi T. Gen Thorac Cardiovasc Surg. 2016 Sep;64(9):517-23. doi: 10.1007/s11748-016-0682-8. Epub 2016 Jul 4. Gen Thorac Cardiovasc Surg. 2016. PMID: 27376535 Review. - Viruses in cancer cell plasticity: the role of hepatitis C virus in hepatocellular carcinoma.
Hibner U, Grégoire D. Hibner U, et al. Contemp Oncol (Pozn). 2015;19(1A):A62-7. doi: 10.5114/wo.2014.47132. Contemp Oncol (Pozn). 2015. PMID: 25691824 Free PMC article. Review.
References
- Ambros IM, Hata J, Joshi VV, Roald B, Dehner LP, Tuchler H, Potschger U, Shimada H. Morphologic features of neuroblastoma (Schwannian stroma-poor tumors) in clinically favorable and unfavorable groups. Cancer. 2002;94:1574–1583. - PubMed
- Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. - PubMed
- Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23:6306–6315. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases