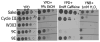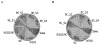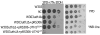Turbidostat culture of Saccharomyces cerevisiae W303-1A under selective pressure elicited by ethanol selects for mutations in SSD1 and UTH1 - PubMed (original) (raw)
Turbidostat culture of Saccharomyces cerevisiae W303-1A under selective pressure elicited by ethanol selects for mutations in SSD1 and UTH1
Liat Avrahami-Moyal et al. FEMS Yeast Res. 2012 Aug.
Abstract
We investigated the genetic causes of ethanol tolerance by characterizing mutations selected in Saccharomyces cerevisiae W303-1A under the selective pressure of ethanol. W303-1A was subjected to three rounds of turbidostat, in a medium supplemented with increasing amounts of ethanol. By the end of selection, the growth rate of the culture has increased from 0.029 to 0.32 h(-1) . Unlike the progenitor strain, all yeast cells isolated from this population were able to form colonies on medium supplemented with 7% ethanol within 6 days, our definition of ethanol tolerance. Several clones selected from all three stages of selection were able to form dense colonies within 2 days on solid medium supplemented with 9% ethanol. We sequenced the whole genomes of six clones and identified mutations responsible for ethanol tolerance. Thirteen additional clones were tested for the presence of similar mutations. In 15 of 19 tolerant clones, the stop codon in ssd1-d was replaced with an amino acid-encoding codon. Three other clones contained one of two mutations in UTH1, and one clone did not contain mutations in either SSD1 or UTH1. We showed that the mutations in SSD1 and UTH1 increased tolerance of the cell wall to zymolyase and conclude that stability of the cell wall is a major factor in increased tolerance to ethanol.
© 2012 Federation of European Microbiological Societies. Published by Blackwell Publishing Ltd. All rights reserved.
Figures
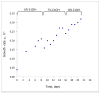
Fig. 1
A significant and rapid increase in the growth rate of a yeast population is observed during selection in a turbidostat. The turbidostat was operated in the range of OD600 1.0-2.0 at 30°C in YPD medium containing ethanol 6% (v/v) for stage I, 7% for stage II and 8% for stage III (see text for details). Growth rate (μ) was measured every 24 hours and the dilution rate was adjusted such that D=μ.
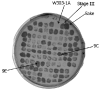
Fig. 2
The culture derived from the selection in turbidostat is heterogeneous. A sample of the turbidostat culture from the last stage of selection was streaked and dispersed on solid YPD medium to obtain single colonies. 105 of these colonies were randomly selected and plated onto solid YPD medium supplemented with 9% ethanol and allowed to grow for 48 hours. The arrows point at the following clones: Sake K9, W303-1A, the mixed population of the last selection stage (Stage III), clone 9E and clone 9C that were selected for further investigation.
Fig. 3
Turbidostat-selected stage III population and clones 9C and 9E are less sensitive to ethanol, caffeine and oxidative stress, compared to parental W303-1A cells. Conditions tested were: 30°C, YPD: 24 h; YPD+9% Ethanol: 72 h; YPD + 8mM Caffeine: 7 days; YNB+ 1mM H2O2: 48h.
Fig. 4
A single gene is responsible for the ability of clones 9C and 9E to proliferate in the presence of high ethanol concentrations. Clones 9E and 9C were mated with W303-1Aα to create diploids W303-1A/9E (Panel A) and W303-1A/9C (Panel B). Following sporulation, the tetrads were dissected. Spores of W303-1A/9E were marked as 9E_S1, 9E_S2, 9E_S3 and 9E_S4. Spores of W303-1A/9C were marked as 9C_S1, 9C_S2, 9C_S3 and 9C_S4. Diploids, as well as resulting pores, were grown on solid YPD+ 7% ethanol. Sake K9 was used as a positive control. The strains were photographed after 4 days in culture at 30°C.
Fig. 5
All stop codon substitutions in the ssd1-d allele confer ethanol tolerance similar to the SSD1-V allele. The ssd1-d alleles with the stop codon substituted for one encoding Trp, Ser, Glu or Gln were constructed by site direct mutagenesis and sub-cloned into pRS416. Aliquots (5μl) of 10-fold serial dilutions of Sake, W303-1A and W303-1A transformed with pRS416 (empty plasmid) or pRS416, which carried the indicated ssd1-d variants, were plated onto YPD +7% ethanol and cultured at 30°C for 5 days. Growth of the same strains under unconstrained conditions (without ethanol) was compared, while strains without plasmids were grown on YPD medium and strains with plasmid were grown on YNB-Ura medium.
Fig. 6
The deletion mutant W303-1A _uth1_Δ proliferates in the presence of ethanol, but less efficiently than clones expressing the UTH1D224Y mutation. Aliquots (5μl) of 10-fold serial dilutions of W303-1A, W303-1A _uth1_Δ, W303-1A _uth1_Δ containing pRS306 vector expressing either UTH1 of W303-1A origin or UTH1D224Y of 9C origin, as well as the clone 9C were plated onto the designated media and cultured at 30°C for 6 days.
Fig. 7
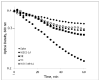
W303-1A_uth1_Δ, 9C and 9E are more tolerant to zymolyase than the parental W303-1A. Cells of the W303-1A_uth1_Δ, 9E, 9C, Sake and W303-1A strains, were allowed to grow on YPD media to stationary growth phase, collected by centrifugation, washed three times with deionized water and then resuspended to an OD600 = 0.4 in TE buffer followed by the addition of zymolyase at the time t = 0. The OD600 of cell suspensions was then recorded at 3 min intervals. Assays were performed in triplicate and were highly reproducible; the standard deviation (SD) did not exceed 10% of the mean value.
Similar articles
- Overexpression of PDE2 or SSD1-V in Saccharomyces cerevisiae W303-1A strain renders it ethanol-tolerant.
Avrahami-Moyal L, Braun S, Engelberg D. Avrahami-Moyal L, et al. FEMS Yeast Res. 2012 Jun;12(4):447-55. doi: 10.1111/j.1567-1364.2012.00795.x. Epub 2012 Mar 29. FEMS Yeast Res. 2012. PMID: 22380741 - Cell morphogenesis proteins are translationally controlled through UTRs by the Ndr/LATS target Ssd1.
Wanless AG, Lin Y, Weiss EL. Wanless AG, et al. PLoS One. 2014 Jan 21;9(1):e85212. doi: 10.1371/journal.pone.0085212. eCollection 2014. PLoS One. 2014. PMID: 24465507 Free PMC article. - SUN family proteins Sun4p, Uth1p and Sim1p are secreted from Saccharomyces cerevisiae and produced dependently on oxygen level.
Kuznetsov E, Kučerová H, Váchová L, Palková Z. Kuznetsov E, et al. PLoS One. 2013 Sep 11;8(9):e73882. doi: 10.1371/journal.pone.0073882. eCollection 2013. PLoS One. 2013. PMID: 24040106 Free PMC article. - Anti-aging effects of hesperidin on Saccharomyces cerevisiae via inhibition of reactive oxygen species and UTH1 gene expression.
Sun K, Xiang L, Ishihara S, Matsuura A, Sakagami Y, Qi J. Sun K, et al. Biosci Biotechnol Biochem. 2012;76(4):640-5. doi: 10.1271/bbb.110535. Epub 2012 Apr 7. Biosci Biotechnol Biochem. 2012. PMID: 22484922 - Ganodermasides A and B, two novel anti-aging ergosterols from spores of a medicinal mushroom Ganoderma lucidum on yeast via UTH1 gene.
Weng Y, Xiang L, Matsuura A, Zhang Y, Huang Q, Qi J. Weng Y, et al. Bioorg Med Chem. 2010 Feb;18(3):999-1002. doi: 10.1016/j.bmc.2009.12.070. Epub 2010 Jan 6. Bioorg Med Chem. 2010. PMID: 20093034
Cited by
- Enhanced Wort Fermentation with De Novo Lager Hybrids Adapted to High-Ethanol Environments.
Krogerus K, Holmström S, Gibson B. Krogerus K, et al. Appl Environ Microbiol. 2018 Jan 31;84(4):e02302-17. doi: 10.1128/AEM.02302-17. Print 2018 Feb 15. Appl Environ Microbiol. 2018. PMID: 29196294 Free PMC article. - Screening and transcriptomic analysis of the ethanol-tolerant mutant Saccharomyces cerevisiae YN81 for high-gravity brewing.
Yang T, Zhang S, Li L, Tian J, Li X, Pan Y. Yang T, et al. Front Microbiol. 2022 Aug 25;13:976321. doi: 10.3389/fmicb.2022.976321. eCollection 2022. Front Microbiol. 2022. PMID: 36090078 Free PMC article. - Adaptation to High Ethanol Reveals Complex Evolutionary Pathways.
Voordeckers K, Kominek J, Das A, Espinosa-Cantú A, De Maeyer D, Arslan A, Van Pee M, van der Zande E, Meert W, Yang Y, Zhu B, Marchal K, DeLuna A, Van Noort V, Jelier R, Verstrepen KJ. Voordeckers K, et al. PLoS Genet. 2015 Nov 6;11(11):e1005635. doi: 10.1371/journal.pgen.1005635. eCollection 2015 Nov. PLoS Genet. 2015. PMID: 26545090 Free PMC article. - From Saccharomyces cerevisiae to Ethanol: Unlocking the Power of Evolutionary Engineering in Metabolic Engineering Applications.
Topaloğlu A, Esen Ö, Turanlı-Yıldız B, Arslan M, Çakar ZP. Topaloğlu A, et al. J Fungi (Basel). 2023 Sep 29;9(10):984. doi: 10.3390/jof9100984. J Fungi (Basel). 2023. PMID: 37888240 Free PMC article. Review. - A novel AST2 mutation generated upon whole-genome transformation of Saccharomyces cerevisiae confers high tolerance to 5-Hydroxymethylfurfural (HMF) and other inhibitors.
Vanmarcke G, Deparis Q, Vanthienen W, Peetermans A, Foulquié-Moreno MR, Thevelein JM. Vanmarcke G, et al. PLoS Genet. 2021 Oct 8;17(10):e1009826. doi: 10.1371/journal.pgen.1009826. eCollection 2021 Oct. PLoS Genet. 2021. PMID: 34624020 Free PMC article.
References
- Alexandre H, Ansanay-Galeote V, Dequin S, Blondin B. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. Febs Letters. 2001;498:98–103. - PubMed
- Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. - PubMed
- Bandara PDS, Flattery-O’Brien JA, Grant CM, Dawes IW. Involvement of the Saccharomyces cerevisiae UTH1 gene in the oxidative-stress response. Current Genetics. 1998;34:259–268. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases