Transcription factor FoxO1, the dominant mediator of muscle wasting in chronic kidney disease, is inhibited by microRNA-486 - PubMed (original) (raw)
Transcription factor FoxO1, the dominant mediator of muscle wasting in chronic kidney disease, is inhibited by microRNA-486
Jing Xu et al. Kidney Int. 2012 Aug.
Abstract
Chronic kidney disease (CKD) accelerates muscle protein degradation by stimulating the ubiquitin proteasome system through activation of the E3 ligases, Atrogin-1/MAFbx and MuRF-1. Forkhead transcription factors (FoxOs) can control the expression of these E3 ligases, but the contribution of individual FoxOs to muscle wasting is unclear. To study this we created mice with a muscle-specific FoxO1 deletion. The absence of FoxO1 blocked 70% of the increase in E3 ligase induction by CKD as well as the proteolysis and loss of muscle mass. Thus, FoxO1 has a role in controlling ubiquitin proteasome system-related proteolysis. As microRNA (miR)-486 reportedly dampens FoxO1 expression and its activity,we transfected a miR-486 mimic into primary cultures of myotubes and found this blocked dexamethasone-stimulated protein degradation without influencing protein synthesis.It also decreased FoxO1 protein translation and increased FoxO1 phosphorylation by downregulation of PTEN phosphatase, a negative regulator of p-Akt. To test its efficacy in vivo, we electroporated miR-486 into muscles and found that the expression of the E3 ligases was suppressed and muscle mass increased despite CKD. Thus, FoxO1 is a dominant mediator of CKD-induced muscle wasting, and miR-486 coordinately decreases FoxO1 and PTEN to protect against this catabolic response.
Figures
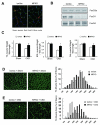
Fig 1. Muscle-specific FoxO1 knockout (MFKO) prevents CKD-induced muscle atrophy
A: FoxO1 protein is absent in myofibers of MFKO mice as assessed by immunostaining. B: Western blot of muscles form MFKO mice revealed that FoxO1 was markedly decreased while FoxO3a and FoxO4 levels were unchanged. C: muscle mass was evaluated by muscle weight normalized to tibia bone length, MFKO prevented the loss of weight of tibialis anterior (TA), extensor digitorum longus (EDL) and soleus (Sol) muscles in CKD mice (*, p<0.05 vs. lox/lox +CKD, n=5). D: The distribution of muscle fiber sizes in control (lox/lox) or MFKO mice was identical (n = 3, >200 myofibers in each mouse were examined). E: The leftward-shift of muscle fiber sizes in lox/lox mice with CKD was prevented in MFKO mice with CKD (n = 5, >200 myofibers in each mouse were examined).
Fig 2. Proteolysis and ubiquitin E3 ligases were largely blocked in muscles of MFKO mice with CKD
A: the absence of FoxO1 minimally influenced the rates of protein synthesis in EDL and Sol muscles (n = 5). B: Rates of protein degradation in EDL and Sol muscles of MFKO and lox/lox mice indicated that deletion of FoxO1 suppressed the increase in muscle proteolysis stimulated by CKD (n = 5 in). C: The expression of Atrogin-1/MAFbx and MuRF-1 mRNAs in TA muscles was accessed by northern blotting. The increased expression of Atrogin-1/MAFbx and MuRF-1 in muscle of CKD, lox/lox mice were eliminated in muscles of MFKO mice with CKD (*, p<0.01 vs. lox/lox+CKD, n = 5). D: Phosphorylation of Akt and FoxOs transcription factors were examined by western blotting. CKD suppressed the p-Akt (Ser 473) and p-FoxO1 (Thr 24) and FoxO3a (Thr 32), but did not change the p-FoxO4 (Ser 262) in muscles of lox/lox and MFKO mice.
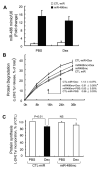
Fig 3. miR-486 blocked Dex-stimulated protein degradation in myotubes
A: the miR-486 mimic (miR-486mc) was transfected into a primary culture of myotubes and the efficiency of transfection was assessed with q-Real time PCR. The miR-486 mimic increased more than 12-fold compared to results in cells transfected with control miRNA (CTL-miR). B: the miR-486 mimic suppressed muscle proteolysis stimulated by Dex. Primary culture of myotubes (transfected with miR-486mimic or CTL-miR) were incubated with [3H] tyrosine overnight and then treated with PBS or 2 m Dex. The released radioactivity (indicating proteins degraded) was plotted as a percentage of total [3H] tyrosine incorporated into cell proteins. The rates of proteolysis (calculated from the linear slopes between 16 and 24 hr) are shown. Measurements were done in duplicate and independently repeated tree times (*, p<0.05 vs. CTL-miR+Dex). C: miR-486 mimic exerted minimal changes in protein synthesis in primary culture of myotubes. Protein synthesis was measured as the incorporation of [3H]-tyrosine after treatment with or without Dex (2 M). Measurements were done in duplicate and independently repeated 3 times.
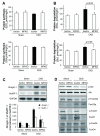
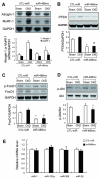
Fig 4. miR-486 suppresses Dex-stimulated Atrogin-1/MAFbx and MuRF-1 expression
A: Atrogin-1/ MAFbx and MuRF-1 expression in primary cultures of myotubes were determined by northern blot. mRNAs of the ubiquitin E3 ligases in response to Dex was blocked by the miR-486mimic (*, p<0.01 vs. CTL-miR+Dex, n=5). B: The levels of p-Akt (ser 473) and PTEN in these myotubes were assessed by western blotting. C: The increase in p-Akt was associated with a decrease in PTEN content in myotubes transfected with miR-486 mimic. D: The miR-486mimic stimulated the phosphorylation of FoxO1 and decreased the FoxO1 protein content. *, p<0.01 vs. CTL-miR+PBS; #, p<0.01 vs. CTL-miR+Dex; n=5.
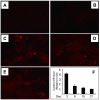
Fig 5. Validation of miRNA electroporation in vivo
TA muscles were injected with Dy547-labeled, control miRNA (cel-miR-67) and the efficiency of transfection was assessed by measuring fluorescence (A to E). A: TA muscles injected with saline or B: injected with Dy547-labeled, control miRNA without electroporation. C: At 2 days after electroporation, Dy547-labeled miRNA was present in myofibers. The fluorescence was detected at 8 and 15 days after electroporation (D and E). F: the cel-miR-67 miRNA levels were examined by RT-PCR.
Fig 6. Enhancing miR-486 protects against CKD-induced loss of skeletal muscle
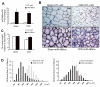
A: TA muscles of sham-operated control, lox/lox (Sham) or CKD mice ( FVB background) were transfected with control miRNA (CTL-miR) or miR-486 mimic (miR-486mc). miR-486 mimic was detected by RT-PCR at 2-weeks after electroporation. B: The cross-sectional area of myofibers in TA muscles following electroporation with the miR-486 mimic or CTL-miR are shown, In situ hybridization revealed that miR-486 was present in myofibers of TA muscles. C: The weight of TA muscles (factored by tibia length) was improved in CKD mice transfected with miR-486mimic (*, p<0.05; n = 5 in each group). D: The distribution of myofiber sizes in muscles from sham and CKD mice treated with CTL-miR or miR-486 mimic. Data were obtained from 7 animals in each group.
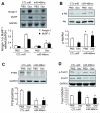
Fig 7. miR-486 suppresses the expression of ubiquitin E3 ligases in muscles of CKD mice
A: The expression of Atrogin-1/MAFbx and MuRF-1 mRNAs was accessed by northern blotting. These E3 ubiquitin ligases were suppressed in muscles of CKD mice treated with miR-486 mimic (*, p<0.01 vs. CTL-miR+CKD; n = 7). B: PTEN was evaluated by western blotting in muscles of sham and CKD mice electroporated with miR-486mimic or with CTL-miR C: FoxO1 was evaluated by western blotting in muscles of sham and CKD mice electroporated with miR-486 mimic or with CTL-miR. Along with an increase in p-Akt, p-FoxO1 levels were raised in muscles electroporated with miR-486 mimic. D: Western blots also reveal an increase in p-Akt in muscles electroporated with miR-486 mimic vs results in muscles electroporated with CTL-miR . *p<0.01 vs. CTL-miR+Sham; #p<0.01 vs. CTL-miR+CKD; n = 5. E: qRT-PCR analysis of miRNAs from muscles of CKD mice electroporated with CTL-miR or miR-486mimic.
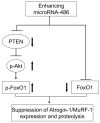
Fig 8. enhancing miR-486 suppresses skeletal muscle proteolysis stimulated by catabolic conditions
In atrophying muscle cells, miR-486 represses the translation of PTEN leading to increased phosphorylation of Akt and FoxO1; miR-486 also directly suppresses FoxO1 translation. These two actions result the inhibition of ubiquitin E3 ligases to block muscle wasting.
Comment in
- MicroRNAs: a new therapeutic frontier for muscle wasting in chronic kidney disease.
Mak RH, Cheung WW. Mak RH, et al. Kidney Int. 2012 Aug;82(4):373-4. doi: 10.1038/ki.2012.150. Kidney Int. 2012. PMID: 22846810
Similar articles
- MicroRNA-23a and MicroRNA-27a Mimic Exercise by Ameliorating CKD-Induced Muscle Atrophy.
Wang B, Zhang C, Zhang A, Cai H, Price SR, Wang XH. Wang B, et al. J Am Soc Nephrol. 2017 Sep;28(9):2631-2640. doi: 10.1681/ASN.2016111213. Epub 2017 Apr 11. J Am Soc Nephrol. 2017. PMID: 28400445 Free PMC article. - Suppression of atrogin-1 and MuRF1 prevents dexamethasone-induced atrophy of cultured myotubes.
Castillero E, Alamdari N, Lecker SH, Hasselgren PO. Castillero E, et al. Metabolism. 2013 Oct;62(10):1495-502. doi: 10.1016/j.metabol.2013.05.018. Epub 2013 Jul 15. Metabolism. 2013. PMID: 23866982 - Excessive glucocorticoid-induced muscle MuRF1 overexpression is independent of Akt/FoXO1 pathway.
Wang XJ, Xiao JJ, Liu L, Jiao HC, Lin H. Wang XJ, et al. Biosci Rep. 2017 Nov 17;37(6):BSR20171056. doi: 10.1042/BSR20171056. Print 2017 Dec 22. Biosci Rep. 2017. PMID: 29046370 Free PMC article. - The role of E3 ubiquitin-ligases MuRF-1 and MAFbx in loss of skeletal muscle mass.
Rom O, Reznick AZ. Rom O, et al. Free Radic Biol Med. 2016 Sep;98:218-230. doi: 10.1016/j.freeradbiomed.2015.12.031. Epub 2015 Dec 29. Free Radic Biol Med. 2016. PMID: 26738803 Review. - The ubiquitin-proteasome system and skeletal muscle wasting.
Attaix D, Ventadour S, Codran A, Béchet D, Taillandier D, Combaret L. Attaix D, et al. Essays Biochem. 2005;41:173-86. doi: 10.1042/EB0410173. Essays Biochem. 2005. PMID: 16250905 Review.
Cited by
- Direct repression of the oncogene CDK4 by the tumor suppressor miR-486-5p in non-small cell lung cancer.
Shao Y, Shen YQ, Li YL, Liang C, Zhang BJ, Lu SD, He YY, Wang P, Sun QL, Jin YX, Ma ZL. Shao Y, et al. Oncotarget. 2016 Jun 7;7(23):34011-21. doi: 10.18632/oncotarget.8514. Oncotarget. 2016. PMID: 27049724 Free PMC article. - Acute Kidney Injury in Septic Patients Treated by Selected Nephrotoxic Antibiotic Agents-Pathophysiology and Biomarkers-A Review.
Petejova N, Martinek A, Zadrazil J, Kanova M, Klementa V, Sigutova R, Kacirova I, Hrabovsky V, Svagera Z, Stejskal D. Petejova N, et al. Int J Mol Sci. 2020 Sep 26;21(19):7115. doi: 10.3390/ijms21197115. Int J Mol Sci. 2020. PMID: 32993185 Free PMC article. Review. - Mechanisms of IGF-1-Mediated Regulation of Skeletal Muscle Hypertrophy and Atrophy.
Yoshida T, Delafontaine P. Yoshida T, et al. Cells. 2020 Aug 26;9(9):1970. doi: 10.3390/cells9091970. Cells. 2020. PMID: 32858949 Free PMC article. Review. - The role of microRNA in cancer cachexia and muscle wasting: A review article.
Sutandyo N. Sutandyo N. Caspian J Intern Med. 2021 Mar;12(2):124-128. doi: 10.22088/cjim.12.2.124. Caspian J Intern Med. 2021. PMID: 34012527 Free PMC article. Review. - Silencing of miR-486 alleviates LPS-stimulated inflammatory response of macrophages through targeting SIRT1.
Huang J, Fu X, Chen X, Xu S, Yu J. Huang J, et al. RSC Adv. 2019 May 31;9(30):17057-17064. doi: 10.1039/c9ra01374a. eCollection 2019 May 29. RSC Adv. 2019. PMID: 35519896 Free PMC article.
References
- Hu Z, Lee IH, Wang X, et al. PTEN expression contributes to the regulation of muscle protein degradation in diabetes. Diabetes. 2007;56:2449–2456. - PubMed
- Lee SW, Dai G, Hu Z, et al. Regulation of muscle protein degradation: coordinated control of apoptotic and ubiquitin-proteasome systems by phosphatidylinositol 3 kinase. J Am Soc Nephrol. 2004;15:1537–1545. - PubMed
- Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–1819. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR060268/AR/NIAMS NIH HHS/United States
- R37 DK037175/DK/NIDDK NIH HHS/United States
- R37 DK037175-29/DK/NIDDK NIH HHS/United States
- R37 DK37175/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous