Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans - PubMed (original) (raw)
. 2012 May 8;109(19):7391-6.
doi: 10.1073/pnas.1202484109. Epub 2012 Apr 18.
Mark S Sharpley, Olga Derbeneva, Leonardo Scherer Alves, Pin Qian, Yaoli Wang, Dimitra Chalkia, Maria Lvova, Jiancheng Xu, Wei Yao, Mariella Simon, Julia Platt, Shiqin Xu, Alessia Angelin, Antonio Davila, Taosheng Huang, Ping H Wang, Lee-Ming Chuang, Lorna G Moore, Guisheng Qian, Douglas C Wallace
Affiliations
- PMID: 22517755
- PMCID: PMC3358837
- DOI: 10.1073/pnas.1202484109
Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans
Fuyun Ji et al. Proc Natl Acad Sci U S A. 2012.
Abstract
The distinction between mild pathogenic mtDNA mutations and population polymorphisms can be ambiguous because both are homoplasmic, alter conserved functions, and correlate with disease. One possible explanation for this ambiguity is that the same variant may have different consequences in different contexts. The NADH dehydrogenase subunit 1 (ND1) nucleotide 3394 T > C (Y30H) variant is such a case. This variant has been associated with Leber hereditary optic neuropathy and it reduces complex I activity and cellular respiration between 7% and 28% on the Asian B4c and F1 haplogroup backgrounds. However, complex I activity between B4c and F1 mtDNAs, which harbor the common 3394T allele, can also differ by 30%. In Asia, the 3394C variant is most commonly associated with the M9 haplogroup, which is rare at low elevations but increases in frequency with elevation to an average of 25% of the Tibetan mtDNAs (odds ratio = 23.7). In high-altitude Tibetan and Indian populations, the 3394C variant occurs on five different macrohaplogroup M haplogroup backgrounds and is enriched on the M9 background in Tibet and the C4a4 background on the Indian Deccan Plateau (odds ratio = 21.9). When present on the M9 background, the 3394C variant is associated with a complex I activity that is equal to or higher than that of the 3394T variant on the B4c and F1 backgrounds. Hence, the 3394C variant can either be deleterious or beneficial depending on its haplogroup and environmental context. Thus, this mtDNA variant fulfills the criteria for a common variant that predisposes to a "complex" disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
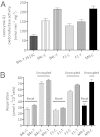
Fig. 1.
Alternations in complex I-specific activity and the cybrid physiology associated with the 3394C variant. (A) NADH: coenzyme q1 oxidoreductase activity of mitochondria isolated from B4c, F1, and M9 haplogroup cybrids. The number of independent determinations for each sample is: B4c-T 3515C (n = 6); B4c-C (n = 19); B4c-T (n = 21); F1-C (n = 5); F1-T (n = 5); M9-C (n = 11). (B) The cellular respiration rates (basal and uncoupled) of the B4c, F1, and M9 cybrid clones. The number of independent determinations for each sample is: B4c-C: (n = 4); B4c-T (n = 6); F1-C (n = 5); F1-T (n = 5); and M9-C (n = 9). On the x axis, the letters next to each haplogroup represent the 3394 variants, C: 3394C and T: 3394T. The error bars represent the SEM.
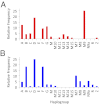
Fig. 2.
Distribution of mtDNA haplogroups in Tibetan and low-land Chinese populations. (A) Tibetan samples 1 and 2, n = 289. (B) SW Chinese, Taiwanese, and southern California Chinese samples, n = 1,644. Haplogroups H, M2, M21, M28, M33, N, U1, and Y with percentages <0.7 in both populations were excluded for visual clarity.
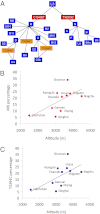
Fig. 3.
Multiple independent origins of the 3394C variant in Tibet. (A) A simplified phylogenetic tree of mtDNA haplogroups and subhaplogroups observed among native Tibetans in sample 1. Steps in which the T3394C variant must have occurred are shown, revealing three independent mutational events. Note that all 3394C variants are associated with macrohaplogroup M, which is also true for the C4a4 mtDNAs of the Deccan plateau, Karnataka State. (B) A positive linear relationship between the frequency of the haplogroup M9 and altitude in Tibetan villages (_R_2 = 0.4018). (C) A positive linear relationship between the frequency of the 3394C variant and altitude in Tibetan villages (_R_2 = 0.3748). The names of the villages sampled are presented next to each symbol and the M9 and 3394C frequencies are provided in
Table S12.
Similar articles
- Leber's Hereditary Optic Neuropathy-Specific Mutation m.11778G>A Exists on Diverse Mitochondrial Haplogroups in India.
Khan NA, Govindaraj P, Soumittra N, Sharma S, Srilekha S, Ambika S, Vanniarajan A, Meena AK, Uppin MS, Sundaram C, Bindu PS, Gayathri N, Taly AB, Thangaraj K. Khan NA, et al. Invest Ophthalmol Vis Sci. 2017 Aug 1;58(10):3923-3930. doi: 10.1167/iovs.16-20695. Invest Ophthalmol Vis Sci. 2017. PMID: 28768321 - Mitochondrial ND1 Variants in 1281 Chinese Subjects With Leber's Hereditary Optic Neuropathy.
Ji Y, Liang M, Zhang J, Zhu L, Zhang Z, Fu R, Liu X, Zhang M, Fu Q, Zhao F, Tong Y, Sun Y, Jiang P, Guan MX. Ji Y, et al. Invest Ophthalmol Vis Sci. 2016 May 1;57(6):2377-89. doi: 10.1167/iovs.16-19243. Invest Ophthalmol Vis Sci. 2016. PMID: 27177320 - Mitochondrial haplogroup background may influence Southeast Asian G11778A Leber hereditary optic neuropathy.
Kaewsutthi S, Phasukkijwatana N, Joyjinda Y, Chuenkongkaew W, Kunhapan B, Tun AW, Suktitipat B, Lertrit P. Kaewsutthi S, et al. Invest Ophthalmol Vis Sci. 2011 Jul 1;52(7):4742-8. doi: 10.1167/iovs.10-5816. Invest Ophthalmol Vis Sci. 2011. PMID: 21398275 - Extranuclear DNA Variations in the Susceptibility of Glaucoma.
Kumar S, Kaur R, Malik MA, Angmo D, Kaur J. Kumar S, et al. Middle East Afr J Ophthalmol. 2024 Jun 14;30(2):113-120. doi: 10.4103/meajo.meajo_132_23. eCollection 2023 Apr-Jun. Middle East Afr J Ophthalmol. 2024. PMID: 39006929 Free PMC article. Review. - [Human mitochondrial genome polymorphism database (mtSNP)].
Fuku N, Nishigaki Y, Tanaka M. Fuku N, et al. Tanpakushitsu Kakusan Koso. 2005 Nov;50(14 Suppl):1753-8. Tanpakushitsu Kakusan Koso. 2005. PMID: 16318308 Review. Japanese. No abstract available.
Cited by
- Genetics: Mitochondrial DNA in evolution and disease.
Wallace DC. Wallace DC. Nature. 2016 Jul 28;535(7613):498-500. doi: 10.1038/nature18902. Epub 2016 Jul 6. Nature. 2016. PMID: 27383787 Free PMC article. No abstract available. - 14th EUNOS Congress: PORTO, PORTUGAL, 16-19 JUNE 2019.
[No authors listed] [No authors listed] Neuroophthalmology. 2019 Jun 7;43(Suppl 1):1-221. doi: 10.1080/01658107.2019.1608780. eCollection 2019 Jun. Neuroophthalmology. 2019. PMID: 31528195 Free PMC article. No abstract available. - Whole Mitochondrial DNA Sequencing Analysis in 47 Han Populations in Southwest China.
Yao L, Xu Z, Wan L. Yao L, et al. Med Sci Monit. 2019 Aug 29;25:6482-6490. doi: 10.12659/MSM.916275. Med Sci Monit. 2019. PMID: 31464266 Free PMC article. - Functional recurrent mutations in the human mitochondrial phylogeny: dual roles in evolution and disease.
Levin L, Zhidkov I, Gurman Y, Hawlena H, Mishmar D. Levin L, et al. Genome Biol Evol. 2013;5(5):876-90. doi: 10.1093/gbe/evt058. Genome Biol Evol. 2013. PMID: 23563965 Free PMC article. - Analysis of functional variants in mitochondrial DNA of Finnish athletes.
Kiiskilä J, Moilanen JS, Kytövuori L, Niemi AK, Majamaa K. Kiiskilä J, et al. BMC Genomics. 2019 Oct 29;20(1):784. doi: 10.1186/s12864-019-6171-6. BMC Genomics. 2019. PMID: 31664900 Free PMC article.
References
- Wallace DC, Lott MT, Procaccio V. Mitochondrial genes in degenerative diseases, cancer and aging. In: Rimoin DL, Connor JM, Pyeritz RE, Korf BR, editors. Emery and Rimoin’s Principles and Practice of Medical Genetics. 5th Ed. Vol 1. Philadelphia: Churchill Livingstone Elsevier; 2007. pp. 194–298.
- Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. - PubMed
- Wallace DC, Ruiz-Pesini E, Mishmar D. mtDNA variation, climatic adaptation, degenerative diseases, and longevity. Cold Spring Harb Symp Quant Biol. 2003;68:479–486. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS21328/NS/NINDS NIH HHS/United States
- HLBI 079647/PHS HHS/United States
- DK73691/DK/NIDDK NIH HHS/United States
- UL1 TR000153/TR/NCATS NIH HHS/United States
- TW001188/TW/FIC NIH HHS/United States
- AG24373/AG/NIA NIH HHS/United States
- TR000153/TR/NCATS NIH HHS/United States
- R01 AG024373/AG/NIA NIH HHS/United States
- R01 DK073691/DK/NIDDK NIH HHS/United States
- R01 NS021328/NS/NINDS NIH HHS/United States
- R01 AG013154/AG/NIA NIH HHS/United States
- AG13154/AG/NIA NIH HHS/United States