Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint - PubMed (original) (raw)
Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint
Lindsey A Shepperd et al. Curr Biol. 2012.
Abstract
The spindle assembly checkpoint (SAC) is the major surveillance system that ensures that sister chromatids do not separate until all chromosomes are correctly bioriented during mitosis. Components of the checkpoint include Mad1, Mad2, Mad3 (BubR1), Bub3, and the kinases Bub1, Mph1 (Mps1), and Aurora B. Checkpoint proteins are recruited to kinetochores when individual kinetochores are not bound to spindle microtubules or not under tension. Kinetochore association of Mad2 causes it to undergo a conformational change, which promotes its association to Mad3 and Cdc20 to form the mitotic checkpoint complex (MCC). The MCC inhibits the anaphase-promoting complex/cyclosome (APC/C) until the checkpoint is satisfied. SAC silencing derepresses Cdc20-APC/C activity. This triggers the polyubiquitination of securin and cyclin, which promotes the dissolution of sister chromatid cohesion and mitotic progression. We, and others, recently showed that association of PP1 to the Spc7/Spc105/KNL1 family of kinetochore proteins is necessary to stabilize microtubule-kinetochore attachments and silence the SAC. We now report that phosphorylation of the conserved MELT motifs in Spc7 by Mph1 (Mps1) recruits Bub1 and Bub3 to the kinetochore and that this is required to maintain the SAC signal.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
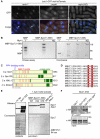
Figure 1. Mph1 kinase phosphorylates MELT motifs in Spc7 to promote association of Spc7 with Bub1
(A) Log phase cultures of bub1-GFP sid4-tdTomato cells either wild type (mph1+), lacking Mph1 (Δmph1) or defective in Mph1 kinase activity (mph1(KD)) were fixed and imaged. Representative images are shown. Cells with mitotic spindles less than 2.5μm exhibiting localised Bub1-GFP (closed arrowheads) or lacking Bub1-GFP localisation (open arrowheads) are highlighted. Bar, 5μm. (B) Mph1 phosphorylates Spc7 in vitro. Mph1 kinase or catalytically inactive Mph1(KD) was incubated with MBP or MBP-Spc7(1-666) fusion protein. Kinase assay (left panel) and coomassie stained gel of input proteins (right panel) are shown. Asterisks indicate Mph1 autophosphorylation. (C) Domain architecture of fission yeast Spc7 and its homologues in budding yeast (S.c. Spc105), worm (C.e. KNL1), and human (H.s. Blinkin). PP1-binding sites (blue), KI motifs (grey), MELT motifs (red) and the coiled-coil kinetochore-binding domain (green) are shown. (D) Protein alignments of the nine MELT motifs in Spc7. Invariant methionine and threonine residues are highlighted in red. (E) Spc7-9TE interacts with Bub1 in vitro. MBP-Spc7 (WT), MBP-Spc7-9TA (9TA), MBP-Spc7-9TE (9TE) or MBP-Pic1-INbox fusion proteins were incubated in extracts of untagged wild type (no tag) or bub1-SZZ cells. Interacting proteins precipitated on amylose beads, separated by SDS-PAGE and subjected to western blot with anti-PAP antibody. Note that the anti-PAP antibody cross-reacts with the MBP-Spc7 proteins. (F) Spc7-9TE interacts with Bub1 in vivo. Extracts from log phase bub1-3HA spc7+, bub1-3HA spc7-GFP or bub1-3HA spc7-9TE-GFP cells were prepared. Proteins were immuno-precipitated with anti-GFP antibodies, separated by SDS-PAGE and subjected to western blot with anti-HA antibodies.
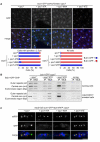
Figure 2. Phosphorylation of the MELT motifs in Spc7 recruits Bub1 to kinetochores
(A) Log phase cultures of Δspc7 spc7+, Δspc7 spc7-9TA, Δspc7 spc7-9MA and Δspc7 spc7-9TE expressing bub1-GFP sid4-tdTomato were fixed. The percentage of pre-anaphase mitotic cells (n=3; left panels) or all cells (n=3; left panels) with Bub1 foci (closed arrowheads, blue bars) or lacking foci (open arrowheads, red bars) was assessed. Bar, 5μm. (B) Log phase or metaphase arrested cultures of Δspc7 spc7+, Δspc7 spc7-9TA and Δspc7 spc7-9TE cells expressing nda3-KM311 bub1-GFP or nda3-KM311 cells (no tag) were fixed and subjected to ChIP analysis with anti-GFP antibodies. The immunoprecipitated DNA was amplified by PCR using primers specific to the central core (cnt), which is the site of kinetochore assembly, and control primers that amplify a noncentromeric, euchromatic negative control (fbp) or centromeric outer repeats (otr). Representative PCRs are shown for each tagged protein, as well as the untagged negative control. (C) Cultures of Δspc7 spc7+, Δspc7 spc7-9TA, Δspc7 spc7-9MA and Δspc7 spc7-9TE expressing bub1-GFP fta3-mRFP pREP3x-mad2 were incubated in medium lacking thiamine for 18 hours to induce Mad2 over-expression. Cells were fixed and imaged. Representative images are shown. Bar, 2μm.
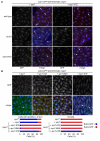
Figure 3. Mph1-dependent recruitment of Bub1 to kinetochores requires Bub3
(A) Log phase cultures of bub1-GFP sid4-tdTomato Δspc7 spc7+ and bub1-GFP sid4-tdTomato Δspc7 spc7-9TE cells either wild type (wild type), lacking Bub3 (Δbub3) or defective in Mph1 kinase activity (mph1(KD)) were fixed and imaged. Cells with mitotic spindles less than 2.5μm exhibiting localised Bub1-GFP (closed arrowheads) or lacking Bub1-GFP localisation (open arrowheads) are highlighted. Bar, 5μm. (B) Log phase cultures of Δspc7 spc7+, Δspc7 spc7-9TA, Δspc7 spc7-9MA and Δspc7 spc7-9TE expressing bub3-GFP sid4-tdTomato were fixed. The percentage of pre-anaphase mitotic cells (n=3; left panels) or all cells (n=3; left panels) with Bub1 foci (closed arrowheads, blue bars) or lacking foci (open arrowheads, red bars) was assessed. Bar, 5μm.
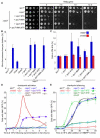
Figure 4. Phosphorylation of Spc7 MELT motifs is required for accurate chromosome segregation and maintenance of the spindle checkpoint
(A) spc7-MELT mutants are sensitive to thiabendazole (TBZ). Serial dilutions of wild type (spc7+), Δspc7 spc7, Δspc7 spc7-9TA, Δspc7 spc7-9MA, Δspc7 spc7-9TE, and Δspc7 spc7-9MA,9TA cells were plated onto YEA plates containing indicated TBZ concentrations and incubated for 3 days at 30°C. (B) Loss of the Ch16(ade6-M216) mini-chromosome was measured using a colony-sectoring assay. Error bars represent the standard deviation of three independent experiments. (C) Anaphase onset is profoundly delayed in spc7-9TA, 9MA and 9MA,9TA mutants but only slightly delayed in spc7-9TE, mutants. Log phase cultures of wild type (spc7+), Δspc7 spc7, Δspc7 spc7-9TA, Δspc7 spc7-9MA, Δspc7 spc7-9TE, and Δspc7 spc7-9MA,9TA cells, expressing cdc13-GFP in the presence (blue bars) or absence (red bars) of Mad3 were fixed and the percentage of cells with Cdc13 on spindles and separated spindle pole bodies was assessed. Error bars represent the standard deviation from three independent experiments. (D) Phosphorylation of Spc7 MELT motifs is required for maintenance of the spindle checkpoint. Log phase cultures of wild type (spc7+), Δmad3, Δbub3, Δspc7 spc7, Δspc7 spc7-9TA and Δspc7 spc7-9TA Δbub3 cells expressing nda3-KM311 ark1-as3 nsk1-GFP were synchronised in early G2 by lactose gradient centrifugation and incubated at 18°C for the times indicated. The cells were fixed and the percentage of cells with spindle pole associated Nsk1 was assessed. (E) Ectopic recruitment of Bub1 and Bub3 to Spc7 aids spindle checkpoint silencing. Log phase cultures of wild type (spc7+), Δdis2, Δbub3, Δspc7 spc7, Δspc7 spc7-9TE and Δspc7 spc7-9TE Δbub3 cells expressing nda3-KM311 ark1-as3 nsk1-GFP were synchronised in prometaphase by incubating at 18°C for 6 hours and 5μM 1NMPP1 was added. At the times indicated the cells were fixed and the percentage of cells with spindle pole associated Nsk1 was assessed. Error bars represent the standard deviation from three independent experiments.
Similar articles
- Bub3-Bub1 Binding to Spc7/KNL1 Toggles the Spindle Checkpoint Switch by Licensing the Interaction of Bub1 with Mad1-Mad2.
Mora-Santos MD, Hervas-Aguilar A, Sewart K, Lancaster TC, Meadows JC, Millar JB. Mora-Santos MD, et al. Curr Biol. 2016 Oct 10;26(19):2642-2650. doi: 10.1016/j.cub.2016.07.040. Epub 2016 Sep 8. Curr Biol. 2016. PMID: 27618268 - The Bub1-TPR Domain Interacts Directly with Mad3 to Generate Robust Spindle Checkpoint Arrest.
Leontiou I, London N, May KM, Ma Y, Grzesiak L, Medina-Pritchard B, Amin P, Jeyaprakash AA, Biggins S, Hardwick KG. Leontiou I, et al. Curr Biol. 2019 Jul 22;29(14):2407-2414.e7. doi: 10.1016/j.cub.2019.06.011. Epub 2019 Jun 27. Curr Biol. 2019. PMID: 31257143 Free PMC article. - MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components.
Yamagishi Y, Yang CH, Tanno Y, Watanabe Y. Yamagishi Y, et al. Nat Cell Biol. 2012 Jun 3;14(7):746-52. doi: 10.1038/ncb2515. Nat Cell Biol. 2012. PMID: 22660415 - The spindle checkpoint: how do cells delay anaphase onset?
Sczaniecka MM, Hardwick KG. Sczaniecka MM, et al. SEB Exp Biol Ser. 2008;59:243-56. SEB Exp Biol Ser. 2008. PMID: 18368927 Review. - Molecular Regulation of the Spindle Assembly Checkpoint by Kinases and Phosphatases.
Manic G, Corradi F, Sistigu A, Siteni S, Vitale I. Manic G, et al. Int Rev Cell Mol Biol. 2017;328:105-161. doi: 10.1016/bs.ircmb.2016.08.004. Epub 2016 Oct 18. Int Rev Cell Mol Biol. 2017. PMID: 28069132 Review.
Cited by
- Borealin dimerization mediates optimal CPC checkpoint function by enhancing localization to centromeres and kinetochores.
Bekier ME, Mazur T, Rashid MS, Taylor WR. Bekier ME, et al. Nat Commun. 2015 Apr 9;6:6775. doi: 10.1038/ncomms7775. Nat Commun. 2015. PMID: 25854549 Free PMC article. - Enrichment of Aurora B kinase at the inner kinetochore controls outer kinetochore assembly.
Bonner MK, Haase J, Swinderman J, Halas H, Miller Jenkins LM, Kelly AE. Bonner MK, et al. J Cell Biol. 2019 Oct 7;218(10):3237-3257. doi: 10.1083/jcb.201901004. Epub 2019 Sep 16. J Cell Biol. 2019. PMID: 31527147 Free PMC article. - Principles and dynamics of spindle assembly checkpoint signalling.
McAinsh AD, Kops GJPL. McAinsh AD, et al. Nat Rev Mol Cell Biol. 2023 Aug;24(8):543-559. doi: 10.1038/s41580-023-00593-z. Epub 2023 Mar 24. Nat Rev Mol Cell Biol. 2023. PMID: 36964313 Review. - Histone H2A phosphorylation recruits topoisomerase IIα to centromeres to safeguard genomic stability.
Zhang M, Liang C, Chen Q, Yan H, Xu J, Zhao H, Yuan X, Liu J, Lin S, Lu W, Wang F. Zhang M, et al. EMBO J. 2020 Feb 3;39(3):e101863. doi: 10.15252/embj.2019101863. Epub 2019 Nov 26. EMBO J. 2020. PMID: 31769059 Free PMC article. - ULK1 phosphorylates Mad1 to regulate spindle assembly checkpoint.
Yuan F, Jin X, Li D, Song Y, Zhang N, Yang X, Wang L, Zhu WG, Tian C, Zhao Y. Yuan F, et al. Nucleic Acids Res. 2019 Sep 5;47(15):8096-8110. doi: 10.1093/nar/gkz602. Nucleic Acids Res. 2019. PMID: 31291454 Free PMC article.
References
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. - PubMed
- Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. - PubMed
- Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 083610/WT_/Wellcome Trust/United Kingdom
- 084229/WT_/Wellcome Trust/United Kingdom
- G0601118/MRC_/Medical Research Council/United Kingdom
- 092076/WT_/Wellcome Trust/United Kingdom
- 077707/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 091020/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous