The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis - PubMed (original) (raw)
The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis
Alexander Knoll et al. Plant Cell. 2012 Apr.
Abstract
The human hereditary disease Fanconi anemia leads to severe symptoms, including developmental defects and breakdown of the hematopoietic system. It is caused by single mutations in the FANC genes, one of which encodes the DNA translocase FANCM (for Fanconi anemia complementation group M), which is required for the repair of DNA interstrand cross-links to ensure replication progression. We identified a homolog of FANCM in Arabidopsis thaliana that is not directly involved in the repair of DNA lesions but suppresses spontaneous somatic homologous recombination via a RecQ helicase (At-RECQ4A)-independent pathway. In addition, it is required for double-strand break-induced homologous recombination. The fertility of At-fancm mutant plants is compromised. Evidence suggests that during meiosis At-FANCM acts as antirecombinase to suppress ectopic recombination-dependent chromosome interactions, but this activity is antagonized by the ZMM pathway to enable the formation of interference-sensitive crossovers and chromosome synapsis. Surprisingly, mutation of At-FANCM overcomes the sterility phenotype of an At-MutS homolog4 mutant by apparently rescuing a proportion of crossover-designated recombination intermediates via a route that is likely At-MMS and UV sensitive81 dependent. However, this is insufficient to ensure the formation of an obligate crossover. Thus, At-FANCM is not only a safeguard for genome stability in somatic cells but is an important factor in the control of meiotic crossover formation.
Figures
Figure 1.
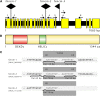
Gene and Protein Structure of At-FANCM. (A) At_-FANCM_ is split into 23 exons and 22 introns with a length of 7085 bp from start to stop codon. T-DNA insertions of mutant lines fancm-1, fancm-2, and fancm-3 were detected in introns 2, 15, and 18, respectively. Large black arrows point in the direction of left border sequences. Small numbered arrows refer to the primers used in genotyping. Primers 7 and 8 were used to assess presence of At_-FANCM_ genomic sequence downstream of the insertion of line fancm-3. (B) The At-FANCM protein has a length of 1344 amino acids (aa) and contains a bipartite helicase domain composed of a DEXDc and a HELICc domain at its N terminus. (C) Detailed analysis of the T-DNA insertion loci. Genomic sequences are shown in bold type, and sequence duplications are underlined. Insertion of foreign sequences is in italic, deletions are named “Del,” and their respective length is given.
Figure 2.
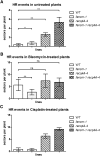
Somatic HR Frequencies. Recombination frequencies in the IC9 recombination reporter background. (A) Spontaneous HR frequencies in untreated plants were slightly but significantly higher in fancm-1 compared with the wild type (WT) (mean 0.715 versus 0.414 sectors per plant). In recq4A-4, the HR frequency was higher still with 2.633 spp. With 5.313 spp, the double mutant fancm-1 recq4A-4 had a HR frequency significantly higher than either single mutant, indicating two parallel pathways of HR suppression. (B) Following induction of DSBs by 5 µg/mL bleomycin, the mean HR frequency of wild-type plants was at 5.62 spp and that of recq4A-4 was not significantly different from the wild type with 7.398 spp. In fancm-1, there was a significant reduction in HR frequency to 2.685 spp compared with the wild type, while the double mutant had 6.8 spp and was not different from the wild type or recq4A-4. (C) Treatment with cisplatin induces DNA ICLs. Induction of HR with 3 µM cisplatin increased the HR rate of wild-type plants to 0.82 spp (compared with 0.414 spp uninduced). Similar to the uninduced assay, the fancm-1 recq4A double mutant exhibits a synergistic increase in HR.
Figure 3.
Representative Meiotic Stages from Pollen Mother Cells. DAPI-stained chromatin spreads of wild-type (WT) ([A], [E], [I], [M], [Q], and [U]), fancm-1 ([B], [F], [J], [N], [R], and [V]), msh4-1 ([C], [G], [K], [O], [S], and [W]), and fancm-1 msh4-1 ([D], [H], [L], [P], [T], and [X]) meiocytes. Metaphase I stages are additionally labeled with 45S (green) and 5S (red) fluorescence in situ hybridization probes to distinguish chromosomes. In pachytene ([A] to [D]), incomplete synapsis, chromatin breaks, and chromosomal interlocks ([B], arrow) are visible in fancm-1 as well as the double mutant. In diakinesis ([E] to [H]), bivalents form in all lines except msh4-1. In fancm-1 and the double mutant, however, connections between bivalents are often detected. Such chromatin bridges are also visible in metaphase I ([I] to [L]) nuclei of fancm-1 ([J], arrow) and fancm-1 msh4-1, leading to unequal distribution of chromosomes in anaphase I ([M] to [P], arrow), which is also visible at the tetrad stage ([U] to [X]). Bars = 10 µm.
Figure 4.
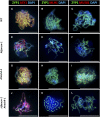
Dual Immunolocalization of Meiotic Proteins ZYP1, ASY1, MLH1, and MUS81 in Pachytene Stage Male Meiocytes. Representative cells of the wild type (WT) ([A] to [C]), fancm-1 ([D] to [F]), msh4-1 ([G] to [I]), and fancm-1 msh4-1 ([J] to [L]) are shown. SC axial element protein ZYP1 is stained green in all cells ([A] to [L]). Detection of SC lateral element ASY1 ([A], [D], [G], and [J]; red) shows incomplete synapsis in fancm-1 and fancm-1 msh4-1 ([D] and [J]). Staining of MLH1 ([B], [E], [H], and [K]) or MUS81 ([C], [F], [I], and [L]) in red shows a reduction of MLH1 foci in msh4-1 and fancm-1 msh4-1 and an increase of MUS81 foci in Atfancm-1 msh4-1. Chromatin is also stained with DAPI. Bars = 10 µm.
Figure 5.
Dual Immunolocalization of Meiotic Proteins ASY1, γH2AX, DMC1, and MSH4 in Pollen Mother Cells. Representative cells of the wild type (WT) ([A] to [C]), fancm-1 ([D] to [F]), msh4-1 ([G] to [I]), and fancm-1 msh4-1 ([J] to [L]) are shown. SC lateral element protein ASY1 is stained green in all cells ([A] to [L]). Numbers of DSB marker γH2AX ([A], [D], [G], and [J]; red) and recombinase DMC1 ([B], [E], [H], and [K]; red) are similar in all lines. MSH4 protein foci ([C], [F], [I], and [L]; red) can be detected in the wild type and fancm-1 in similar quantities but not in msh4-1 or the double mutant. Chromatin is also stained with DAPI. Bars = 10 µm.
Figure 6.
Epistasis Analysis of Meiosis Phenotypes. Chromatin spreads of double mutant fancm-1 spo11-2-3, fancm-1 rad51, and fancm-1 rmi1-1 as well as the respective single mutant and wild-type (WT) meiocytes. Shown are informative meiotic stages. (A) and (B) Diakinesis meiocytes of the wild type and fancm-1 are paired into five bivalents (A), while there are 10 univalents visible in corresponding meiocytes of spo11-2-3 as well as the double mutant line fancm-1 spo11-2-3 (B). In rad51 mutant meiocytes, SPO11-induced DSBs cannot be repaired, which results in a random segregation of chromatin fragments seen in tetrads. Similarly, in the double mutant fancm-1 rad51, chromatin fragmentation is detectable, but not in fancm-1 or wild-type meiocytes. (C) In the wild type and fancm-1 anaphase I, homologous chromosomes are pulled toward opposite poles. In rmi1-1, there are unresolved recombination intermediates between homologous chromosomes that result in chromosome bridges and fragmentation. Similar defects can be found in the double mutant fancm-1 rmi1-1. Bar = 10 µm.
Figure 7.
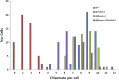
Distribution of Chiasmata Numbers per Cell in the Wild Type, msh4-1, fancm-1, and fancm-1 msh4-1. Chiasmata numbers were counted in metaphase I cells stained with DAPI as well as 5S and 45S fluorescence in situ hybridization probes in the wild type (WT; blue), msh4-1 (red), fancm-1 (green), and the double mutant fancm-1 msh4-1 (purple). The number of chiasmata was slightly reduced in fancm-1 compared with the wild type. The strongly reduced chiasmata count of msh4-1 could be partially rescued by further mutation of At_-FANCM_, as can be seen in the respective double mutant. Interestingly, in fancm-1, univalents were never observed, although as few as six chiasmata were counted in some cells, indicating that an obligate CO per chromosome is maintained.
Similar articles
- Fanconi anemia ortholog FANCM regulates meiotic crossover distribution in plants.
Li X, Yu M, Bolaños-Villegas P, Zhang J, Ni D, Ma H, Wang Y. Li X, et al. Plant Physiol. 2021 May 27;186(1):344-360. doi: 10.1093/plphys/kiab061. Plant Physiol. 2021. PMID: 33576801 Free PMC article. - CDKG1 Is Required for Meiotic and Somatic Recombination Intermediate Processing in Arabidopsis.
Nibau C, Lloyd A, Dadarou D, Betekhtin A, Tsilimigka F, Phillips DW, Doonan JH. Nibau C, et al. Plant Cell. 2020 Apr;32(4):1308-1322. doi: 10.1105/tpc.19.00942. Epub 2020 Feb 10. Plant Cell. 2020. PMID: 32047050 Free PMC article. - The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants.
Knoll A, Puchta H. Knoll A, et al. J Exp Bot. 2011 Mar;62(5):1565-79. doi: 10.1093/jxb/erq357. Epub 2010 Nov 16. J Exp Bot. 2011. PMID: 21081662 Review. - A tumor suppressive DNA translocase named FANCM.
Basbous J, Constantinou A. Basbous J, et al. Crit Rev Biochem Mol Biol. 2019 Feb;54(1):27-40. doi: 10.1080/10409238.2019.1568963. Epub 2019 Feb 4. Crit Rev Biochem Mol Biol. 2019. PMID: 30714416 Review.
Cited by
- The nuclease FAN1 is involved in DNA crosslink repair in Arabidopsis thaliana independently of the nuclease MUS81.
Herrmann NJ, Knoll A, Puchta H. Herrmann NJ, et al. Nucleic Acids Res. 2015 Apr 20;43(7):3653-66. doi: 10.1093/nar/gkv208. Epub 2015 Mar 16. Nucleic Acids Res. 2015. PMID: 25779053 Free PMC article. - Why do plants need the ZMM crossover pathway? A snapshot of meiotic recombination from the perspective of interhomolog polymorphism.
Ziolkowski PA. Ziolkowski PA. Plant Reprod. 2023 Mar;36(1):43-54. doi: 10.1007/s00497-022-00446-3. Epub 2022 Jul 12. Plant Reprod. 2023. PMID: 35819509 Free PMC article. - High-Resolution Mapping of Crossover Events in the Hexaploid Wheat Genome Suggests a Universal Recombination Mechanism.
Darrier B, Rimbert H, Balfourier F, Pingault L, Josselin AA, Servin B, Navarro J, Choulet F, Paux E, Sourdille P. Darrier B, et al. Genetics. 2017 Jul;206(3):1373-1388. doi: 10.1534/genetics.116.196014. Epub 2017 May 22. Genetics. 2017. PMID: 28533438 Free PMC article. - Unravelling mechanisms that govern meiotic crossover formation in wheat.
Higgins JD, Osman K, Desjardins SD, Henderson IR, Edwards KJ, Franklin FCH. Higgins JD, et al. Biochem Soc Trans. 2022 Aug 31;50(4):1179-1186. doi: 10.1042/BST20220405. Biochem Soc Trans. 2022. PMID: 35901450 Free PMC article. Review. - Fancm has dual roles in the limiting of meiotic crossovers and germ cell maintenance in mammals.
Tsui V, Lyu R, Novakovic S, Stringer JM, Dunleavy JEM, Granger E, Semple T, Leichter A, Martelotto LG, Merriner DJ, Liu R, McNeill L, Zerafa N, Hoffmann ER, O'Bryan MK, Hutt K, Deans AJ, Heierhorst J, McCarthy DJ, Crismani W. Tsui V, et al. Cell Genom. 2023 Jun 29;3(8):100349. doi: 10.1016/j.xgen.2023.100349. eCollection 2023 Aug 9. Cell Genom. 2023. PMID: 37601968 Free PMC article.
References
- Allers T., Lichten M. (2001). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57 - PubMed
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
- Armstrong S.J., Caryl A.P., Jones G.H., Franklin F.C. (2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115: 3645–3655 - PubMed
- Armstrong S.J., Sanchez-Moran E., Franklin F.C. (2009). Cytological analysis of Arabidopsis thaliana meiotic chromosomes. In Methods in Molecular Biology: Meiosis: Cytological Methods, Vol. 2, S. Keeney, ed (New York: Humana Press; Springer), pp. 131–145. - PubMed
- Bakker S.T., van de Vrugt H.J., Rooimans M.A., Oostra A.B., Steltenpool J., Delzenne-Goette E., van der Wal A., van der Valk M., Joenje H., te Riele H., de Winter J.P. (2009). Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum. Mol. Genet. 18: 3484–3495 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases