Genetic structure and demographic history should inform conservation: Chinese cobras currently treated as homogenous show population divergence - PubMed (original) (raw)
Genetic structure and demographic history should inform conservation: Chinese cobras currently treated as homogenous show population divergence
Long-Hui Lin et al. PLoS One. 2012.
Abstract
An understanding of population structure and genetic diversity is crucial for wildlife conservation and for determining the integrity of wildlife populations. The vulnerable Chinese cobra (Naja atra) has a distribution from the mouth of the Yangtze River down to northern Vietnam and Laos, within which several large mountain ranges and water bodies may influence population structure. We combined 12 microsatellite loci and 1117 bp of the mitochondrial cytochrome b gene to explore genetic structure and demographic history in this species, using 269 individuals from various localities in Mainland China and Vietnam. High levels of genetic variation were identified for both mtDNA and microsatellites. mtDNA data revealed two main (Vietnam + southern China + southwestern China; eastern + southeastern China) and one minor (comprising only two individuals from the westernmost site) clades. Microsatellite data divided the eastern + southeastern China clade further into two genetic clusters, which include individuals from the eastern and southeastern regions, respectively. The Luoxiao and Nanling Mountains may be important barriers affecting the diversification of lineages. In the haplotype network of cytchrome b, many haplotypes were represented within a "star" cluster and this and other tests suggest recent expansion. However, microsatellite analyses did not yield strong evidence for a recent bottleneck for any population or genetic cluster. The three main clusters identified here should be considered as independent management units for conservation purposes. The release of Chinese cobras into the wild should cease unless their origin can be determined, and this will avoid problems arising from unnatural homogenization.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
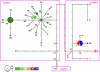
Figure 1. Three genetic clusters of Naja atra in China and Vietnam based on the admixture model in Structure.
Individuals are represented by a point which is colored to reflect the cluster to which it was assigned with the highest membership coefficient. Large points represent individuals with membership coefficient _q_≥0.90 and small ones q<0.90. Ellipses show the three main genetic clusters. See Table 1 for sample site abbreviations.
Figure 2. Network of 39 mitochondrial cytochrome b haplotypes from 269 individual Naja atra.
The size of the circles is proportional to haplotype frequency; small open circles represent intermediate haplotypes that are not in our study. See Table 1 for sample sites abbreviations. Sites where most individuals were assigned to the same cluster on the basis of microsatellite data have gradations of the same color (s).
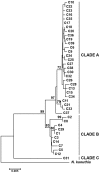
Figure 3. Maximum-likelihood tree for all 39 haplotypes of Naja atra and for one outgroup taxon.
Labels are haplotype identification numbers. Values above branches indicate support for each node based on maximum likelihood. Bootstrap values below 70% are not shown.
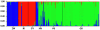
Figure 4. Admixture analysis of individual genotypes using Structure.
Colors correspond to one of three clusters (see also Fig. 1), and each bar represents a single sample. See Table 1 for sample site abbreviations.
Similar articles
- The phylogeographical pattern and conservation of the Chinese cobra (Naja atra) across its range based on mitochondrial control region sequences.
Lin LH, Hua L, Qu YF, Gao JF, Ji X. Lin LH, et al. PLoS One. 2014 Sep 3;9(9):e106944. doi: 10.1371/journal.pone.0106944. eCollection 2014. PLoS One. 2014. PMID: 25184236 Free PMC article. - Conservation genetics of the Chinese cobra (Naja atra) investigated with mitochondrial DNA sequences.
Lin LH, Zhao Q, Ji X. Lin LH, et al. Zoolog Sci. 2008 Sep;25(9):888-93. doi: 10.2108/zsj.25.888. Zoolog Sci. 2008. PMID: 19267597 - Phylogeography, speciation and demographic history: Contrasting evidence from mitochondrial and nuclear markers of the Odorrana graminea sensu lato (Anura, Ranidae) in China.
Chen Z, Li H, Zhai X, Zhu Y, He Y, Wang Q, Li Z, Jiang J, Xiong R, Chen X. Chen Z, et al. Mol Phylogenet Evol. 2020 Mar;144:106701. doi: 10.1016/j.ympev.2019.106701. Epub 2019 Dec 4. Mol Phylogenet Evol. 2020. PMID: 31811937 - Preliminary insights into the phylogeography of the yellow-bellied sea snake, Pelamis platurus.
Sheehy CM 3rd, Solórzano A, Pfaller JB, Lillywhite HB. Sheehy CM 3rd, et al. Integr Comp Biol. 2012 Aug;52(2):321-30. doi: 10.1093/icb/ics088. Epub 2012 Jun 1. Integr Comp Biol. 2012. PMID: 22659201 - Isolation and characterization of microsatellite loci in the Chinese Cobra Naja atra (Elapidae).
Lin LH, Mao LX, Luo X, Qu YF, Ji X. Lin LH, et al. Int J Mol Sci. 2011;12(7):4435-40. doi: 10.3390/ijms12074435. Epub 2011 Jul 7. Int J Mol Sci. 2011. PMID: 21845087 Free PMC article.
Cited by
- Climate warming will increase chances of hybridization and introgression between two Takydromus lizards (Lacertidae).
Guo K, Zhong J, Xie F, Zhu L, Qu YF, Ji X. Guo K, et al. Ecol Evol. 2021 May 11;11(13):8573-8584. doi: 10.1002/ece3.7671. eCollection 2021 Jul. Ecol Evol. 2021. PMID: 34257917 Free PMC article. - The Phylogeography and Population Demography of the Yunnan Caecilian (Ichthyophis bannanicus): Massive Rivers as Barriers to Gene Flow.
Wang H, Luo X, Meng S, Bei Y, Song T, Meng T, Li G, Zhang B. Wang H, et al. PLoS One. 2015 Apr 27;10(4):e0125770. doi: 10.1371/journal.pone.0125770. eCollection 2015. PLoS One. 2015. PMID: 25915933 Free PMC article. - The phylogeny, phylogeography, and diversification history of the westernmost Asian cobra (Serpentes: Elapidae: Naja oxiana) in the Trans-Caspian region.
Kazemi E, Nazarizadeh M, Fatemizadeh F, Khani A, Kaboli M. Kazemi E, et al. Ecol Evol. 2020 Dec 22;11(5):2024-2039. doi: 10.1002/ece3.7144. eCollection 2021 Mar. Ecol Evol. 2020. PMID: 33717439 Free PMC article. - Description of a New Cobra (Naja Laurenti, 1768; Squamata, Elapidae) from China with Designation of a Neotype for Naja atra.
Shi SC, Vogel G, Ding L, Rao DQ, Liu S, Zhang L, Wu ZJ, Chen ZN. Shi SC, et al. Animals (Basel). 2022 Dec 9;12(24):3481. doi: 10.3390/ani12243481. Animals (Basel). 2022. PMID: 36552401 Free PMC article. - Relative contributions of neutral and non-neutral genetic differentiation to inform conservation of steelhead trout across highly variable landscapes.
Matala AP, Ackerman MW, Campbell MR, Narum SR. Matala AP, et al. Evol Appl. 2014 Jun;7(6):682-701. doi: 10.1111/eva.12174. Epub 2014 Jun 27. Evol Appl. 2014. PMID: 25067950 Free PMC article.
References
- Wallach V, Wüster W, Broadley DG. In praise of subgenera: taxonomic status of cobras of the genus Naja Laurenti (Serpentes: Elapidae). Zootaxa. 2009;2236:26–36.
- Wüster W, Crookes S, Ineich I, Mané Y, Pook CE, et al. The phylogeny of cobras inferred from mitochondrial DNA sequences: evolution of venom spitting and the phylogeography of the African spitting cobras (Serpentes: Elapidae: Naja nigricollis complex). Mol Phylogenet Evol. 2007;45:437–453. - PubMed
- Wüster W, Thorpe RS. Asiatic cobras: population systematics of the Naja naja species complex (Serpentes: Elapidae) in India and Central Asia. Herpetologica. 1992;48:69–85.
- Szyndlar Z, Rage JC. West Palearctic cobras of the genus Naja (Serpentes: Elapidae): interrelationships among extinct and extant species. Amphibia-Reptilia. 1990;11:385–400.
- Ineich I. Etat actuel de nos connaissances sur la classification des serpents venimeux. Bull Soc Herpétol France 1995. 1995;75–76:7–24.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources