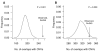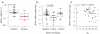Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents - PubMed (original) (raw)
doi: 10.1158/2159-8290.CD-11-0206. Epub 2012 Mar 22.
Zhigang C Wang # 2 3, Ji-Young Kim 3 4, Aron C Eklund 1, Qiyuan Li 1 2, Ruiyang Tian 2, Christian Bowman-Colin 2, Yang Li 2, April Greene-Colozzi 2, J Dirk Iglehart 2 3, Nadine Tung 5, Paula D Ryan 6, Judy E Garber 2, Daniel P Silver # 2 3, Zoltan Szallasi # 1 7, Andrea L Richardson # 2 3
Affiliations
- PMID: 22576213
- PMCID: PMC3806629
- DOI: 10.1158/2159-8290.CD-11-0206
Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents
Nicolai J Birkbak et al. Cancer Discov. 2012 Apr.
Erratum in
- Cancer Discov. 2013 Aug;3(8):952
Abstract
DNA repair competency is one determinant of sensitivity to certain chemotherapy drugs, such as cisplatin. Cancer cells with intact DNA repair can avoid the accumulation of genome damage during growth and also can repair platinum-induced DNA damage. We sought genomic signatures indicative of defective DNA repair in cell lines and tumors and correlated these signatures to platinum sensitivity. The number of subchromosomal regions with allelic imbalance extending to the telomere (N(tAI)) predicted cisplatin sensitivity in vitro and pathologic response to preoperative cisplatin treatment in patients with triple-negative breast cancer (TNBC). In serous ovarian cancer treated with platinum-based chemotherapy, higher levels of N(tAI) forecast a better initial response. We found an inverse relationship between BRCA1 expression and N(tAI) in sporadic TNBC and serous ovarian cancers without BRCA1 or BRCA2 mutation. Thus, accumulation of telomeric allelic imbalance is a marker of platinum sensitivity and suggests impaired DNA repair.
Significance: Mutations in BRCA genes cause defects in DNA repair that predict sensitivity to DNA damaging agents, including platinum; however, some patients without BRCA mutations also benefit from these agents. NtAI, a genomic measure of unfaithfully repaired DNA, may identify cancer patients likely to benefit from treatments targeting defective DNA repair.
Figures
Figure 1. Chromosomal aberrations and cisplatin sensitivity in vitro
The relationship between NtAI and cisplatin sensitivity was analyzed in breast cancer cell lines. A and B: 10 cell lines were included in this study, 1: CAMA-1, 2: HCC1954, 3:MDA-MB-231, 4: MDA-MB-361, 5: HCC1187, 6:BT-549, 7: HCC1143, 8: MDA-MB-468, 9: BT-20, 10: T47D. A. IC50 values for each of the 10 cell lines. A proliferation assay was used to assess viability after 48 hours of cisplatin exposure, and IC50 was determined from the dose response curves. B. Relationship between NtAI and cisplatin sensitivity. Breast cancer subtype is indicated as follows: ER− HER2−, red; HER2+, green, ER+ HER2−, blue. C. Relationship between NtAI and cisplatin sensitivity as determined by GI50 in breast cancer cell lines from Heiser et al. (18). Reported transcriptional subtype is indicated as follows: basal, red; claudin-low, pink; ERBB2Amp, green; luminal, blue. See supplemental methods for cell line identifiers.
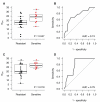
Figure 2. NtAI and cisplatin response in breast cancer
In two clinical trials, TNBC patients were given preoperative cisplatin (Cisplatin-1, Fig. 2 A-B) or cisplatin and bevacizumab (Cisplatin-2, Fig. 2 C-D). Cisplatin resistant tumors are indicated in black, cisplatin sensitive tumors are indicated in red. Tumors with germline mutations in BRCA1/2 are indicated with triangles. A and C. Box plots showing NtAI distribution in cisplatin resistant and sensitive tumors. B and D. Receiver operating characteristic curves showing the ability of NtAI to predict for sensitivity to cisplatin.
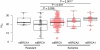
Figure 3. NtAI and cisplatin response in serous ovarian cancer
Box plots showing NtAI distribution in platinum sensitive and resistant tumors in cancers without BRCA1 or BRCA2 mutations (wtBRCA) and for cancers with germline or somatic mutation in BRCA1 (mBRCA1) or in BRCA2 (mBRCA2). Red indicate sensitive samples, triangles indicate samples with germline or somatic mutations in BRCA1 or BRCA2. Significant differences between resistant wtBRCA and sensitive groups are indicated. In addition, significant differences were found between sensitive wtBRCA and sensitive mBRCA2 (P = 0.047), and sensitive wtBRCA and sensitive mBRCA1 (P = 0.014).
Figure 4. Enrichment of common CNVs in tAI chromosomal breakpoints from TNBC
Association of tAI breakpoints with common CNV loci based on computational simulations that compared the expected number of breakpoints containing CNVs with the observed number in total cases in Cisplatin-1 (A) and Cisplatin-2 (B).
Figure 5. Association between BRCA1 transcript level and cisplatin sensitivity, BRCA1 promoter methylation, and NtAI
Red indicates tumors sensitive to cisplatin. Tumors with a germline mutation in BRCA1 or BRCA2 are excluded in A. and B., but included in C., represented as triangles. In B. and C., BRCA1 transcript levels measured by qPCR were scaled and combined by centering the values, and dividing by the standard deviation within each trial. A. BRCA1 transcripts in resistant and sensitive tumors in the Cisplatin-2 cohort. B. BRCA1 expression in tumors by methylation status of the BRCA1 promoter region in the combined Cisplatin-1 and Cisplatin-2 cohorts. C. Relationship of BRCA1 transcript level and NtAI in the combined Cisplatin-1 and Cisplatin-2 cohorts.
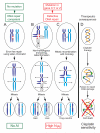
Figure 6. A model relating DNA repair to accumulation of telomeric AI and response to platinum agents
A. In DNA repair-competent cells, DNA breaks are repaired using error-free homologous recombination employing the identical sister chromatid as a template, resulting in no AI. B. and C. Compromised DNA repair favors the use of error-prone repair pathways, resulting in chromosome rearrangements and aberrant radial chromosome formation. After mitotic division, daughter cells will have imbalance in the parental contribution of telomeric segments of chromosomes (telomeric AI). B. Non-homologous end joining is one error-prone mechanism that joins a broken chromatid of one chromosome (dark blue) to the chromatid of another, usually nonhomologous, chromosome (white). Mitotic segregation results in cells with telomeric AI due to mono-allelic change in DNA copy number of the affected telomeric region. C. Mitotic recombination may result in rearrangements between homologous chromosomes (dark blue and light blue). Mitotic segregation results in cells with AI due to copy neutral loss of heterozygosity (LOH). D. The same compromise in DNA repair that causes telomeric AI may also result in the inability of the tumor cell to repair drug-induced DNA damage, leading to tumor sensitivity to drugs such as platinum salts.
Similar articles
- Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer.
Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC, Szallasi Z, Barry WT, Winer EP, Tung NM, Isakoff SJ, Ryan PD, Greene-Colozzi A, Gutin A, Sangale Z, Iliev D, Neff C, Abkevich V, Jones JT, Lanchbury JS, Hartman AR, Garber JE, Ford JM, Silver DP, Richardson AL. Telli ML, et al. Clin Cancer Res. 2016 Aug 1;22(15):3764-73. doi: 10.1158/1078-0432.CCR-15-2477. Epub 2016 Mar 8. Clin Cancer Res. 2016. PMID: 26957554 Free PMC article. Clinical Trial. - Association of BRCA1/2 defects with genomic scores predictive of DNA damage repair deficiency among breast cancer subtypes.
Timms KM, Abkevich V, Hughes E, Neff C, Reid J, Morris B, Kalva S, Potter J, Tran TV, Chen J, Iliev D, Sangale Z, Tikishvili E, Perry M, Zharkikh A, Gutin A, Lanchbury JS. Timms KM, et al. Breast Cancer Res. 2014 Dec 5;16(6):475. doi: 10.1186/s13058-014-0475-x. Breast Cancer Res. 2014. PMID: 25475740 Free PMC article. - Differential contributory roles of nucleotide excision and homologous recombination repair for enhancing cisplatin sensitivity in human ovarian cancer cells.
Wang QE, Milum K, Han C, Huang YW, Wani G, Thomale J, Wani AA. Wang QE, et al. Mol Cancer. 2011 Mar 8;10:24. doi: 10.1186/1476-4598-10-24. Mol Cancer. 2011. PMID: 21385444 Free PMC article. - Targeting DNA repair and replication stress in the treatment of ovarian cancer.
Murai J. Murai J. Int J Clin Oncol. 2017 Aug;22(4):619-628. doi: 10.1007/s10147-017-1145-7. Epub 2017 Jun 22. Int J Clin Oncol. 2017. PMID: 28643177 Review. - [Abnormalities of DNA repair and gynecological cancers].
Auguste A, Leary A. Auguste A, et al. Bull Cancer. 2017 Nov;104(11):971-980. doi: 10.1016/j.bulcan.2017.09.007. Epub 2017 Oct 18. Bull Cancer. 2017. PMID: 29054544 Review. French.
Cited by
- Unlocking the potential of immunotherapy in platinum-resistant ovarian cancer: rationale, challenges, and novel strategies.
Kefas J, Flynn M. Kefas J, et al. Cancer Drug Resist. 2024 Oct 15;7:39. doi: 10.20517/cdr.2024.67. eCollection 2024. Cancer Drug Resist. 2024. PMID: 39534871 Free PMC article. Review. - Development of a prognostic model related to homologous recombination deficiency in glioma based on multiple machine learning.
Gong Z, Zhou D, Shen H, Ma C, Wu D, Hou L, Wang H, Xu T. Gong Z, et al. Front Immunol. 2024 Oct 7;15:1452097. doi: 10.3389/fimmu.2024.1452097. eCollection 2024. Front Immunol. 2024. PMID: 39434883 Free PMC article. - Identification of three subtypes of ovarian cancer and construction of prognostic models based on immune-related genes.
Gao W, Yuan H, Yin S, Deng R, Ji Z. Gao W, et al. J Ovarian Res. 2024 Oct 21;17(1):208. doi: 10.1186/s13048-024-01526-w. J Ovarian Res. 2024. PMID: 39434163 Free PMC article. - Prediction of homologous recombination deficiency from routine histology with attention-based multiple instance learning in nine different tumor types.
Loeffler CML, El Nahhas OSM, Muti HS, Carrero ZI, Seibel T, van Treeck M, Cifci D, Gustav M, Bretz K, Gaisa NT, Lehmann KV, Leary A, Selenica P, Reis-Filho JS, Ortiz-Bruechle N, Kather JN. Loeffler CML, et al. BMC Biol. 2024 Oct 8;22(1):225. doi: 10.1186/s12915-024-02022-9. BMC Biol. 2024. PMID: 39379982 Free PMC article. - DNA damage response in breast cancer and its significant role in guiding novel precise therapies.
Li J, Jia Z, Dong L, Cao H, Huang Y, Xu H, Xie Z, Jiang Y, Wang X, Liu J. Li J, et al. Biomark Res. 2024 Sep 27;12(1):111. doi: 10.1186/s40364-024-00653-2. Biomark Res. 2024. PMID: 39334297 Free PMC article. Review.
References
- Samouelian V, Maugard CM, Jolicoeur M, Bertrand R, Arcand SL, Tonin PN, et al. Chemosensitivity and radiosensitivity profiles of four new human epithelial ovarian cancer cell lines exhibiting genetic alterations in BRCA2, TGFbeta-RII, KRAS2, TP53 and/or CDNK2A. Cancer Chemother Pharmacol. 2004;54:497–504. - PubMed
- Byrski T, Huzarski T, Dent R, Gronwald J, Zuziak D, Cybulski C, et al. Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res Treat. 2009;115:359–363. - PubMed
- Cass I, Baldwin RL, Varkey T, Moslehi R, Narod SA, Karlan BY. Improved survival in women with BRCA-associated ovarian carcinoma. Cancer. 2003;97:2187–2195. - PubMed
- Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–2132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous