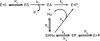Protein engineering of penicillin acylase - PubMed (original) (raw)
Protein engineering of penicillin acylase
V I Tishkov et al. Acta Naturae. 2010 Jul.
Abstract
Penicillin acylases (PA) are widely used for the production of semi-synthetic β-lactam antibiotics and chiral compounds. In this review, the latest achievements in the production of recombinant enzymes are discussed, as well as the results of PA type G protein engineering.
Keywords: E.coli; expression; penicillin acylase; protein engineering; structure.
Figures
Fig. 1
Modern classification of β-lactam acylases according to the type of substrate [7]. 7-ACA – 7-aminocephalosporanic acid
Fig. 2
Alignment of amino acid sequences of penicillin acylases G from different sources. Catalytic βSer1 residue and residues from the oxyanion hole are marked by “*” and “#”, respectively. Sequences of the signal peptide and intersubunit spacer are underlined in bold italic, and underlined, respectively.
Fig. 3
Phylogenetic tree of penicillin acylases G.
Fig. 4
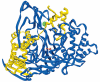
Structure of the active heterodimer of PA-G from E.coli (pdb 1pnk) [24]. α- and β-subunits are shown in yellow and dark blue, respectively. Catalytic βSer1 residue and Ca 2+ ion are shown in red and green, respectively.
Fig. 5
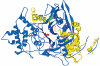
Structure of the active heterodimer of PA-G from A .faecalis (pdb 3K3W). α- and β-subunits are shown in yellow and dark blue, respectively. Catalytic βSer1 residue and Ca 2+ ion are shown in red and green, respectively. Disulfide bond in β-subunit between residues Cys492 and Cys525 in wild-type enzyme is shown in magenta. Insert in right part of figure shows fixation of N-terminus of α-subunit and C-terminus of β-subunit due to creation of new disulfide bond (shown in orange) after double mutation αQ3C/βP751C.
Fig. 6
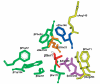
Main amino acid residues in the active site of PA from E. coli (structure PDB 1AI4). Catalytic βSer1 residue and residue βGln23 are shown in red and orange, respectively. Residues from oxyanion hole are presented in magenta. Residues from substrate-binding domain are shown in green and yellow for subdomains S1 and S2, respectively. Residue αPhe146 belonging to both subdomains is in blue.
Scheme. 1
A minimal scheme for half-synthetic β-lactam antibiotics preparation by nucleophilic substitution. E, S, and ES are the enzyme, substrate (donor of acyl moiety), and the enzyme-substrate complex, respectively. EA, Nu, and EANu are the acyl-enzyme, nucleophile, and the double complex of acyl and nucleophile. K s , K n, and K p - are binding constants of substrate free enzyme E, the nucleophile with acyl-enzyme EA, and the product with free enzyme E, respectively.
Similar articles
- Molecular biology of β-lactam acylases.
Deshpande BS, Ambedkar SS, Sudhakaran VK, Shewale JG. Deshpande BS, et al. World J Microbiol Biotechnol. 1994 Mar;10(2):129-38. doi: 10.1007/BF00360873. World J Microbiol Biotechnol. 1994. PMID: 24420933 - Biotechnological advances on penicillin G acylase: pharmaceutical implications, unique expression mechanism and production strategies.
Srirangan K, Orr V, Akawi L, Westbrook A, Moo-Young M, Chou CP. Srirangan K, et al. Biotechnol Adv. 2013 Dec;31(8):1319-32. doi: 10.1016/j.biotechadv.2013.05.006. Epub 2013 May 27. Biotechnol Adv. 2013. PMID: 23721991 Review. - Penicillin G acylase production by Mucor griseocyanus and the partial genetic analysis of its pga gene.
Cano-Cabrera JC, Palomo-Ligas L, Flores-Gallegos AC, Martínez-Hernández JL, Rodríguez-Herrera R. Cano-Cabrera JC, et al. Int Microbiol. 2021 Jan;24(1):37-45. doi: 10.1007/s10123-020-00137-x. Epub 2020 Jul 23. Int Microbiol. 2021. PMID: 32705496 - Saturation mutagenesis reveals the importance of residues alphaR145 and alphaF146 of penicillin acylase in the synthesis of beta-lactam antibiotics.
Jager SA, Shapovalova IV, Jekel PA, Alkema WB, Svedas VK, Janssen DB. Jager SA, et al. J Biotechnol. 2008 Jan 1;133(1):18-26. doi: 10.1016/j.jbiotec.2007.08.039. Epub 2007 Aug 30. J Biotechnol. 2008. PMID: 17933411 - Exploitation of E. coli for the production of penicillin G amidase: a tool for the synthesis of semisynthetic β-lactam antibiotics.
Sambyal K, Singh RV. Sambyal K, et al. J Genet Eng Biotechnol. 2021 Oct 15;19(1):156. doi: 10.1186/s43141-021-00263-7. J Genet Eng Biotechnol. 2021. PMID: 34652570 Free PMC article. Review.
Cited by
- Exploring the Structurally Conserved Regions and Functional Significance in Bacterial N-Terminal Nucleophile (Ntn) Amide-Hydrolases.
Quiroga I, Hernández-González JA, Bautista-Rodríguez E, Benítez-Rojas AC. Quiroga I, et al. Int J Mol Sci. 2024 Jun 21;25(13):6850. doi: 10.3390/ijms25136850. Int J Mol Sci. 2024. PMID: 38999960 Free PMC article. - Strategies to Improve the Biosynthesis of β-Lactam Antibiotics by Penicillin G Acylase: Progress and Prospects.
Pan X, Xu L, Li Y, Wu S, Wu Y, Wei W. Pan X, et al. Front Bioeng Biotechnol. 2022 Jul 18;10:936487. doi: 10.3389/fbioe.2022.936487. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35923572 Free PMC article. Review. - Insight into the Lifestyle of Amoeba Willaertia magna during Bioreactor Growth Using Transcriptomics and Proteomics.
Hasni I, Decloquement P, Demanèche S, Mameri RM, Abbe O, Colson P, La Scola B. Hasni I, et al. Microorganisms. 2020 May 21;8(5):771. doi: 10.3390/microorganisms8050771. Microorganisms. 2020. PMID: 32455615 Free PMC article. - Functional characterization of bacteria isolated from ancient arctic soil exposes diverse resistance mechanisms to modern antibiotics.
Perron GG, Whyte L, Turnbaugh PJ, Goordial J, Hanage WP, Dantas G, Desai MM. Perron GG, et al. PLoS One. 2015 Mar 25;10(3):e0069533. doi: 10.1371/journal.pone.0069533. eCollection 2015. PLoS One. 2015. PMID: 25807523 Free PMC article. - To a Question on the Mechanism of the Antimicrobial Action of Ortho-Benzoic Sulfimide.
Kasap EY, Grishin DV. Kasap EY, et al. Pharmaceuticals (Basel). 2020 Dec 13;13(12):461. doi: 10.3390/ph13120461. Pharmaceuticals (Basel). 2020. PMID: 33322230 Free PMC article. Review.
References
- Sakaguchi K., Murao S.. J. Agr. Chem. S. 1950;23:1–3.
- Claridge C.A., Luttinger J.R., Lein J.. Proc. Soc. Exp. Biol. Med. 1963;113:1008–1012. - PubMed
- Valle F., Balbas P., Merino E.. Trends Biochem. Sci. 1991;16:36–40. - PubMed
- Arroyo M., de la Mata I., Acebal C.. Appl. Microbiol. Biotechnol. 2003;60:507–514. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources