Cyclic di-GMP sensing via the innate immune signaling protein STING - PubMed (original) (raw)
Cyclic di-GMP sensing via the innate immune signaling protein STING
Qian Yin et al. Mol Cell. 2012.
Abstract
Detection of foreign materials is the first step of successful immune responses. Stimulator of interferon genes (STING) was shown to directly bind cyclic diguanylate monophosphate (c-di-GMP), a bacterial second messenger, and to elicit strong interferon responses. Here we elucidate the structural features of the cytosolic c-di-GMP binding domain (CBD) of STING and its complex with c-di-GMP. The CBD exhibits an α + β fold and is a dimer in the crystal and in solution. Surprisingly, one c-di-GMP molecule binds to the central crevice of a STING dimer, using a series of stacking and hydrogen bonding interactions. We show that STING is autoinhibited by an intramolecular interaction between the CBD and the C-terminal tail (CTT) and that c-di-GMP releases STING from this autoinhibition by displacing the CTT. The structures provide a remarkable example of pathogen-host interactions in which a unique microbial molecule directly engages the innate immune system.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
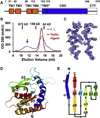
Figure 1. Overall Structure of the STING c-di-GMP Binding Domain
(A) Domain organization of STING. Five previously suggested transmembrane helices (TMs) are represented with four orange boxes (TM1–4) and a blue box (TM5), followed by the suggested cytosolic domain. The c-di-GMP binding domain (CBD) identified from this study, which contains the previous TM5, is represented by the blue box and the long blue rectangle. The construct used in E. coli expression (139–379) is represented with the dotted box. CTT: C-terminal tail. Starting residue number of each domain is labeled above. (B) Gel filtration profiles of full-length (F.L., blue) and trypsin-digested (red) cytosolic domain. Elution volumes of protein standards are marked at the top. The black arrow points to high molecular weight species observed only in fulllength cytosolic domain. (C) Experimental electron density of the α1–α1′ (left) and β4–α3 (right) region contoured at 1.4 σ. (D) Cartoon representation of one STING protomer. The molecule is in rainbow spectrum with blue at the N terminus and red at the C terminus. N and C termini as well as secondary structure elements are labeled. Disordered regions are drawn as dotted lines. (E) Topology diagram of the STING CBD. α helices and β strands are represented by cylinders and wide arrows, respectively. Peptide directionality is indicated by black arrows. The molecule is colored in rainbow spectrum as in (D).
Figure 2. Structural Features of the STING CBD
Secondary structure elements are labeled on top of the human STING sequence. α helices are shown as green cylinders,β strands as red arrows, and loops as yellow lines. Residues without defined densities are shown as dotted yellow lines. Solvent accessibility is shown underneath each residue by a blue-white gradient. Buried residues are indicated by blue shadings and exposed residues by white shadings. Residues of STING conserved among different species (Figure S1) are highlighted in yellow. Residues at the dimer interface are colored in green, and c-di-GMP contacting residues are in red. Residue T267, involved both in dimerization and c-di-GMP binding, is colored in magenta.
Figure 3. Dimeric Nature of STING CBD
(A) Cartoon representation of STING dimer. One protomer is colored as in Figure 2 with α helices in green, β strands in red, and loops in yellow. The other protomer is colored in gray. The interacting α1 and α3 are labeled for both protomers. (B) Detailed interaction at the dimer interface. For clarity, only one of the two symmetrical interfaces is shown. One molecule is colored in green and the other in gray, as in (A). Interfacial residues are shown as sticks. Residues and the helices they belong to are labeled and numbered. (C) The STING dimer is rotated roughly 90° horizontally from the orientation in (A) and colored in the same way. The three hydrophobic cores are labeled. Residues constituting the hydrophobic cores are shown as sticks. Note core 3 is composed of residues from both protomers. (D) Surface representation of the STING dimer in a side view, showing the deep crevice across the dimer interface, and the electrostatic surface of the STING dimer shown in a top view.
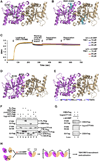
Figure 4. Recognition of c-di-GMP by STING
(A) Difference Fourier (Fo-Fc) map contoured at 2.5 σ at the dimer interface, superimposed with the bound c-di-GMP. (B) Cartoon representation of STING/c-di-GMP complex. One STING molecule is colored in violet and the other in wheat. The c-di-GMP molecule is shown as a stick model. (C) Superposition of the STING/c-di-GMP complex with the free STING dimer. The STING protomers in the complex are colored in violet and wheat, while those in the free STING are in green and gray. Red arrow marks the slight outward swing of the STING molecule (wheat) in the complex with c-di-GMP. (D) Isothermal titration calorimetry measurement for the interaction between STING and c-di-GMP. (E) Stick representation of c-di-GMP with the intrinsic two-fold symmetry. The orientation is the same as in (C). 2-position nitrogen of the guanine ring, 2′-hydroxyl group of the ribose, and the phosphoryl oxygen atoms are labeled. (F) Detailed interaction of c-di-GMP with STING. For clarity, only one “half site” is shown. Contacting residues are labeled, and hydrogen bonds are represented by dotted lines.
Figure 5. Proposed Mechanism of STING Activation
(A) Mutants that contain buried residues and fail to interact with c-di-GMP. These buried residues, I200, D216, Y245, and Y314, are shown as sticks on a cartoon representation of the STING/c-di-GMP structure. (B) A multiply substituted mutant that contains exposed residues and fails to interact with c-di-GMP. These residues,R281–K289,are coloredincyanand shownassticks. (C) Real time biolayer interferometry measurement of 10, 20, and 40 µM TBK1 to immobilized STING CBD in the absence (black, blue, and purple lines) or presence (green, red, and yellow lines) of c-di-GMP. Biotinylated STING CBD loading and washing as well as TBK1 association and dissociation curves are labeled. (D) Mutants that bind c-di-GMP, hyperinduce IFN but do not respond to c-di-GMP for further enhancement of IFN production. These residues, E316 and S272, are shown as sticks on the STING/c-di-GMP cartoon representation. (E) Mutants in the CTT that bind c-di-GMP, induce IFN but do not respond to c-di-GMP for further enhancement of IFN production. These residues, S358/E360/S366 and R375 are highlighted in blue in the appended CTT. (F) Interaction of CTT with full-length (FL) cytosolic domain of STING and CBD were detected by immunoprecipitation (IP) followed by immunoblotting (IB) in 293 cells transfected with indicated expression constructs. (G) The interaction between CTT and CBD as detected by coimmunoprecipitation is weakened when cells are stimulated with c-di-GMP. (H) “Release of autoinhibition” model. In unstimulated cells, STING exists in an autoinhibited status with an intramolecular CBD (violet or wheat ovals)/CTT (black curvy lines) interaction, either as a monomer or dimer. Binding of c-di-GMP at the dimer interface displaces CTT, induces and stabilizes STING dimer, and releases STING from autoinhibition. STING dimers with freed CTT may further oligomerize through CTT. Activated STING then recruits and activates TBK1 and IRF3.
Similar articles
- Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system.
Shu C, Yi G, Watts T, Kao CC, Li P. Shu C, et al. Nat Struct Mol Biol. 2012 Jun 24;19(7):722-4. doi: 10.1038/nsmb.2331. Nat Struct Mol Biol. 2012. PMID: 22728658 Free PMC article. - Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING.
Chin KH, Tu ZL, Su YC, Yu YJ, Chen HC, Lo YC, Chen CP, Barber GN, Chuah ML, Liang ZX, Chou SH. Chin KH, et al. Acta Crystallogr D Biol Crystallogr. 2013 Mar;69(Pt 3):352-66. doi: 10.1107/S0907444912047269. Epub 2013 Feb 16. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 23519410 - Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding.
Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, Li Y, Zhang R, Cheng G, Liu ZJ. Ouyang S, et al. Immunity. 2012 Jun 29;36(6):1073-86. doi: 10.1016/j.immuni.2012.03.019. Epub 2012 May 10. Immunity. 2012. PMID: 22579474 Free PMC article. - Binding of bacterial secondary messenger molecule c di-GMP is a STING operation.
Shaw N, Ouyang S, Liu ZJ. Shaw N, et al. Protein Cell. 2013 Feb;4(2):117-29. doi: 10.1007/s13238-012-2071-0. Epub 2012 Dec 20. Protein Cell. 2013. PMID: 23264039 Free PMC article. Review. - Cyclic di-nucleotide signaling enters the eukaryote domain.
Schaap P. Schaap P. IUBMB Life. 2013 Nov;65(11):897-903. doi: 10.1002/iub.1212. Epub 2013 Oct 17. IUBMB Life. 2013. PMID: 24136904 Free PMC article. Review.
Cited by
- STING regulates BCR signaling in normal and malignant B cells.
Tang CA, Lee AC, Chang S, Xu Q, Shao A, Lo Y, Spalek WT, Pinilla-Ibarz JA, Del Valle JR, Hu CA. Tang CA, et al. Cell Mol Immunol. 2021 Apr;18(4):1016-1031. doi: 10.1038/s41423-020-00552-0. Epub 2020 Sep 30. Cell Mol Immunol. 2021. PMID: 32999453 Free PMC article. - STING and the innate immune response to nucleic acids in the cytosol.
Burdette DL, Vance RE. Burdette DL, et al. Nat Immunol. 2013 Jan;14(1):19-26. doi: 10.1038/ni.2491. Nat Immunol. 2013. PMID: 23238760 Review. - Structure-function analysis of STING activation by c[G(2',5')pA(3',5')p] and targeting by antiviral DMXAA.
Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L, Hartmann G, Barchet W, Tuschl T, Patel DJ. Gao P, et al. Cell. 2013 Aug 15;154(4):748-62. doi: 10.1016/j.cell.2013.07.023. Epub 2013 Aug 1. Cell. 2013. PMID: 23910378 Free PMC article. - STING modulators: Predictive significance in drug discovery.
Cui X, Zhang R, Cen S, Zhou J. Cui X, et al. Eur J Med Chem. 2019 Nov 15;182:111591. doi: 10.1016/j.ejmech.2019.111591. Epub 2019 Aug 8. Eur J Med Chem. 2019. PMID: 31419779 Free PMC article. Review. - Crystal structure and functional implication of bacterial STING.
Ko TP, Wang YC, Yang CS, Hou MH, Chen CJ, Chiu YF, Chen Y. Ko TP, et al. Nat Commun. 2022 Jan 10;13(1):26. doi: 10.1038/s41467-021-26583-3. Nat Commun. 2022. PMID: 35013136 Free PMC article.
References
- Akira S, Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. - PubMed
- Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459:1015–1018. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials