Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic - PubMed (original) (raw)
Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic
Rui Xu et al. J Virol. 2012 Sep.
Abstract
The 2009 H1N1 influenza pandemic is the first human pandemic in decades and was of swine origin. Although swine are believed to be an intermediate host in the emergence of new human influenza viruses, there is still little known about the host barriers that keep swine influenza viruses from entering the human population. We surveyed swine progenitors and human viruses from the 2009 pandemic and measured the activities of the hemagglutinin (HA) and neuraminidase (NA), which are the two viral surface proteins that interact with host glycan receptors. A functional balance of these two activities (HA binding and NA cleavage) is found in human viruses but not in the swine progenitors. The human 2009 H1N1 pandemic virus exhibited both low HA avidity for glycan receptors as a result of mutations near the receptor binding site and weak NA enzymatic activity. Thus, a functional match between the hemagglutinin and neuraminidase appears to be necessary for efficient transmission between humans and may be an indicator of the pandemic potential of zoonotic viruses.
Figures
Fig 1
List of glycans on the microarray. The glycans include 34 unique natural sialoside epitopes (α2-3 linkage, numbers 3 to 25; α2-6 linkage, numbers 26 to 34; mixed linkage, numbers 35 and 36) that are relevant to influenza biology and two neutral glycans (numbers 1 and 2) as controls.
Fig 2
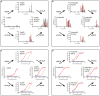
Avidity shift in the HA from swine to human. The avidity shift in the HA is mainly mediated by two changes in the receptor binding site: A227E and R133AK. The swine virus signature residues Ala227 and Arg133A, when substituted into Cali09 HA, improve binding for glycan receptors (A and C), whereas human virus residues Glu227 and Lys133A attenuate receptor binding in sw/Indiana00 HA (B and D). The amino acid substitutions change binding avidity but not binding specificity for glycan linkages as both wild-type strains and mutants maintain favorable binding for α2-6-linked sialic acids. In glycan microarray binding (A and B), binding signals are shown in filled bars colored by sialic acid linkages: neutral glycans are in black (numbers 1 and 2), α2-3-linked glycans in are blue (3 to 25), α2-6-linked glycans in red (26 to 34), and mixed glycans in are cyan (35 and 36). Glycan array results for Cali09 and sw/Indiana00 are shown on different scales. In the plate binding assay (C and D), binding for 6′-SLNLN is shown in red, and that for 3′-SLNLN is in blue. In each panel, wild-type binding of 6′-SLNLN is shown as a dashed line for comparison.
Fig 3
Differences in glycan binding avidity and receptor binding site structure between sw/Indiana00 and Cali09. (A) Comparison of receptor binding by ELISA. The sw/Indiana00 is the HA progenitor of pandemic H1N1 virus Cali09. HAs from both strains display specificity for α2-6-linked sialic acids, but swine HA demonstrates significantly stronger binding toward glycan 6′-SLNLN in a plate-based ELISA. The difference in binding avidities is correlated to structural differences in the HAs (B and C). (B) Crystal structure of the Cali09 HA in complex with the α2-6 receptor analog LSTc (PDB 3UBE) (52). (C) Crystal structure of the sw/Indiana00 HA. (D) Three residue changes are found in the receptor binding site between swine progenitors and the 2009 H1N1 pandemic strains. Two of these residues are in proximity to bound glycan receptors in Cali09: Glu227 and Lys133A (highlighted in red circles in panels B and C). The third residue, Ile219, is located at the periphery of the binding pocket (highlighted in a dashed red circle).
Fig 4
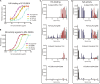
Comparison of HA and NA activities for pandemic viruses. In human pandemic viruses, HA binding to glycan receptors is closely correlated with the efficiency of NA cleavage of these glycan receptors, suggesting that a functional match of these two surface glycoproteins contributes to effective human transmission. Strong HA glycan binding in Japan57 and HK68 is paired up with NAs that exhibit higher activity. Weaker binding in SC18 and Cali09 is complemented by NAs of lower activity. HA binding was measured against biotinylated 6′-SLNLN in a plate assay (A) and printed glycans on a microarray (C) as previously described (52). NA activity was quantified by a solution assay using the fluorescence substrate 4-MU-NANA (B). NA specificity for glycan structures was investigated by the newly developed microarray method (D). All of the NA microarray results shown in the main figures were obtained at 0.8 μg/ml. Results with other NA concentrations are presented in Fig S5 to S14 in the supplemental material.
Fig 5
HA and NA activities of swine H1 viruses. Swine viruses that are closely related to the 2009 H1N1 pandemic virus show strong HA glycan binding (A and C) but variable NA activities (B and D). HA binding was studied by ELISA (A) and on the glycan microarray (C). NA activity was measured by a solution-based assay using fluorescent substrate 4-MU-NANA (B) and by the NA glycan array activity/specificity assay (D). sw/Indiana00 N2 NA shows very weak NA activity. Iowa05 N1 NA is slightly more efficient and is roughly comparable to the Cali09 N1 NA. sw/Guangxi06 N2 NA is very efficient in catalyzing cleavage of 4-MU-NANA and comparable to Japan57 N2 NA. The mismatched Iowa05 virus was isolated from an infected patient but did not cause further human infections.
Fig 6
Functional balance of HA and NA in human pandemic viruses. Human pandemic viruses that readily transmit among humans (red filled circles) exhibit a functional balance of HA receptor binding and NA receptor cleavage activities. The strength of glycan binding in HA is correlated with the efficiency of NA cleavage of glycan receptors. A maladapted human virus (Neth602, red open circles) deviates from such functional balance. Swine progenitors (black filled squares) have strong HA binding but variable levels of NA activity. (A) HA binding avidity plotted against NA activity as expressed in _k_cat/Km. (B) HA binding avidity plotted against NA activity as expressed in _k_cat (the turnover rate under optimum conditions). SC1918, A/South Carolina/1/18.
Fig 7
Adaptive fine-tuning of the 2009 pandemic virus. (A) Amino acid substitutions in HA and NA in two strains that were isolated 4 weeks later than the first isolated Cali09 viruses. The incidence of an amino acid occurring at certain position among 2009 pandemic viruses is shown in parentheses. A total of 2,271 H1 HA sequences and 1,147 N1 NA sequences from 2009 pandemic viruses were available in the Influenza Virus Resource at the NCBI in September 2011 (2). (B and D) The amino acid changes in the HA improved binding for glycan receptors. The NA of strain NY06 also shows improved catalytic efficiency (C and E), corresponding to an increase in both HA and NA activities. Descendants of an NY06-like virus became dominant later in the pandemic. On the other hand, Neth602 displays diminished NA activity (C and E) and an HA/NA imbalance, but the NA mutations acquired by Neth602 did not propagate in later pandemic strains.
Similar articles
- Positive selection on hemagglutinin and neuraminidase genes of H1N1 influenza viruses.
Li W, Shi W, Qiao H, Ho SY, Luo A, Zhang Y, Zhu C. Li W, et al. Virol J. 2011 Apr 21;8:183. doi: 10.1186/1743-422X-8-183. Virol J. 2011. PMID: 21507270 Free PMC article. - Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets.
Yen HL, Liang CH, Wu CY, Forrest HL, Ferguson A, Choy KT, Jones J, Wong DD, Cheung PP, Hsu CH, Li OT, Yuen KM, Chan RW, Poon LL, Chan MC, Nicholls JM, Krauss S, Wong CH, Guan Y, Webster RG, Webby RJ, Peiris M. Yen HL, et al. Proc Natl Acad Sci U S A. 2011 Aug 23;108(34):14264-9. doi: 10.1073/pnas.1111000108. Epub 2011 Aug 8. Proc Natl Acad Sci U S A. 2011. PMID: 21825167 Free PMC article. - Triple-reassortant influenza A virus with H3 of human seasonal origin, NA of swine origin, and internal A(H1N1) pandemic 2009 genes is established in Danish pigs.
Krog JS, Hjulsager CK, Larsen MA, Larsen LE. Krog JS, et al. Influenza Other Respir Viruses. 2017 May;11(3):298-303. doi: 10.1111/irv.12451. Epub 2017 Mar 21. Influenza Other Respir Viruses. 2017. PMID: 28245096 Free PMC article. - Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1).
Brockwell-Staats C, Webster RG, Webby RJ. Brockwell-Staats C, et al. Influenza Other Respir Viruses. 2009 Sep;3(5):207-13. doi: 10.1111/j.1750-2659.2009.00096.x. Influenza Other Respir Viruses. 2009. PMID: 19768134 Free PMC article. Review. - What adaptive changes in hemagglutinin and neuraminidase are necessary for emergence of pandemic influenza virus from its avian precursor?
Gambaryan AS, Matrosovich MN. Gambaryan AS, et al. Biochemistry (Mosc). 2015 Jul;80(7):872-80. doi: 10.1134/S000629791507007X. Biochemistry (Mosc). 2015. PMID: 26542001 Review.
Cited by
- Structural restrictions for influenza neuraminidase activity promote adaptation and diversification.
Wang H, Dou D, Östbye H, Revol R, Daniels R. Wang H, et al. Nat Microbiol. 2019 Dec;4(12):2565-2577. doi: 10.1038/s41564-019-0537-z. Epub 2019 Aug 26. Nat Microbiol. 2019. PMID: 31451775 - A Single Amino Acid Substitution at Residue 218 of Hemagglutinin Improves the Growth of Influenza A(H7N9) Candidate Vaccine Viruses.
Li X, Gao Y, Ye Z. Li X, et al. J Virol. 2019 Sep 12;93(19):e00570-19. doi: 10.1128/JVI.00570-19. Print 2019 Oct 1. J Virol. 2019. PMID: 31270231 Free PMC article. - Phylogenetic analysis and clinical characteristics of the co-occurring mutations in HA and NA genes of influenza A(H1N1)pdm09 viruses during 2015-2017 in Beijing, China.
Liu Y, Wang Y, Liu B, Cong X, Ji Y, Guo X, Gao Y. Liu Y, et al. Virol J. 2020 Nov 19;17(1):182. doi: 10.1186/s12985-020-01446-3. Virol J. 2020. PMID: 33213486 Free PMC article. - The homologous tripartite viral RNA polymerase of A/swine/Korea/CT1204/2009(H1N2) influenza virus synergistically drives efficient replication and promotes respiratory droplet transmission in ferrets.
Pascua PN, Song MS, Kwon HI, Lim GJ, Kim EH, Park SJ, Lee OJ, Kim CJ, Webby RJ, Webster RG, Choi YK. Pascua PN, et al. J Virol. 2013 Oct;87(19):10552-62. doi: 10.1128/JVI.01333-13. Epub 2013 Jul 17. J Virol. 2013. PMID: 23864624 Free PMC article. - Adaptation potential of H3N8 canine influenza virus in human respiratory cells.
Sekine W, Kamiki H, Ishida H, Matsugo H, Ohira K, Li K, Katayama M, Takenaka-Uema A, Murakami S, Horimoto T. Sekine W, et al. Sci Rep. 2024 Aug 13;14(1):18750. doi: 10.1038/s41598-024-69509-x. Sci Rep. 2024. PMID: 39138310 Free PMC article.
References
- Adams PD, et al. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 - PubMed
- Beigel JH, et al. 2005. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med. 353:1374–1385 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical