Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system - PubMed (original) (raw)
Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system
Chang Shu et al. Nat Struct Mol Biol. 2012.
Abstract
STING (stimulator of interferon genes) is an innate immune sensor of cyclic dinucleotides that regulates the induction of type I interferons. STING's C-terminal domain forms a V-shaped dimer and binds a cyclic diguanylate monophosphate (c-di-GMP) at the dimer interface by both direct and solvent-mediated hydrogen bonds. Guanines of c-di-GMP stack against the phenolic rings of a conserved tyrosine, and mutations at the c-di-GMP binding surface reduce nucleotide binding and affect signaling.
Figures
Figure 1
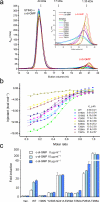
STING forms a dimer and binds c-di-GMP at the dimer interface. (a) C-di-GMP binding studies of human STING CTD (residues 149–341) by gel filtration chromatography. (b) C-di-GMP binding studies of STING CTD (residues 149 to 341) by ITC. (c) C-di-GMP binding studies of STING (residues 155 to 379) by ITC. (d) Structure of human STING CTD bound to c-di-GMP. STING is shown by the red and cyan ribbon representation. C-di-GMP is shown by the yellow ball and stick model.
Figure 2
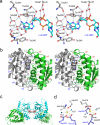
Structural basis of c-di-GMP recognition and the dimer interface of STING. (a) Stereo close up of the c-di-GMP binding site of STING. Residues of STING are shown by the gray ball and stick models. C-di-GMP is shown by the cyan ball and stick model. Solvent molecules mediating hydrogen bonds between STING and c-di-GMP are shown by the red spheres. (b). Stereo close up view of the STING dimer interface. Residues at the dimer interface are shown by the gray and green stick models. (c) The Ca2+ binding sites between two STING dimers. Ca2+ ions are shown by the gray spheres and Ca2+ ligands are shown as gray sticks. Solvent molecules are shown by the red spheres. (d) Close up of the Ca2+ binding sites. Residues from one STING are shown by the gray ball and stick models. Residues from a symmetry-related STING are shown by the purple ball and stick models. Ca2+ are shown by the yellow spheres. Solvent molecules coordinating with the Ca2+ are shown by the blue spheres.
Figure 3
Mutations at the nucleotide binding surface reduce c-di-GMP binding and affect STING signaling. (a) C-di-GMP binding studies of wild-type and mutants of STING CTD by gel filtration chromatography. The inset shows a close up of the unbound c-di-GMP peaks. (b) C-di-GMP binding studies of STING mutants by ITC. (c) IFN-β luciferase assays show that mutations at the c-di-GMP binding surface affect signaling of mouse STING expressed in HEK293T cells. Residues Ile199, Tyr239, Asn241, Glu259, Thr262, Pro263, and Thr266 of mouse STING correspond to residues Ile200, Tyr240, Asn242, Glu260, Thr263, Pro264, and Thr267 of human STING. The error bars show standard deviations of signals from three independent transfections.
Similar articles
- The structural basis for the sensing and binding of cyclic di-GMP by STING.
Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. Huang YH, et al. Nat Struct Mol Biol. 2012 Jun 24;19(7):728-30. doi: 10.1038/nsmb.2333. Nat Struct Mol Biol. 2012. PMID: 22728659 - Cyclic di-GMP sensing via the innate immune signaling protein STING.
Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Yin Q, et al. Mol Cell. 2012 Jun 29;46(6):735-45. doi: 10.1016/j.molcel.2012.05.029. Epub 2012 Jun 14. Mol Cell. 2012. PMID: 22705373 Free PMC article. - STING is a direct innate immune sensor of cyclic di-GMP.
Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. Burdette DL, et al. Nature. 2011 Sep 25;478(7370):515-8. doi: 10.1038/nature10429. Nature. 2011. PMID: 21947006 Free PMC article. - Binding of bacterial secondary messenger molecule c di-GMP is a STING operation.
Shaw N, Ouyang S, Liu ZJ. Shaw N, et al. Protein Cell. 2013 Feb;4(2):117-29. doi: 10.1007/s13238-012-2071-0. Epub 2012 Dec 20. Protein Cell. 2013. PMID: 23264039 Free PMC article. Review. - Targeting c-di-GMP Signaling, Biofilm Formation, and Bacterial Motility with Small Molecules.
Opoku-Temeng C, Sintim HO. Opoku-Temeng C, et al. Methods Mol Biol. 2017;1657:419-430. doi: 10.1007/978-1-4939-7240-1_31. Methods Mol Biol. 2017. PMID: 28889311 Review.
Cited by
- Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, Tuschl T, Patel DJ. Gao P, et al. Cell. 2013 May 23;153(5):1094-107. doi: 10.1016/j.cell.2013.04.046. Epub 2013 May 3. Cell. 2013. PMID: 23647843 Free PMC article. - ICA69 aggravates ferroptosis causing septic cardiac dysfunction via STING trafficking.
Kong C, Ni X, Wang Y, Zhang A, Zhang Y, Lin F, Li S, Lv Y, Zhu J, Yao X, Dai Q, Mo Y, Wang J. Kong C, et al. Cell Death Discov. 2022 Apr 9;8(1):187. doi: 10.1038/s41420-022-00957-y. Cell Death Discov. 2022. PMID: 35397620 Free PMC article. - An Analysis of the Expression and Association with Immune Cell Infiltration of the cGAS/STING Pathway in Pan-Cancer.
An X, Zhu Y, Zheng T, Wang G, Zhang M, Li J, Ji H, Li S, Yang S, Xu D, Li Z, Wang T, He Y, Zhang L, Yang W, Zhao R, Hao D, Li X. An X, et al. Mol Ther Nucleic Acids. 2019 Mar 1;14:80-89. doi: 10.1016/j.omtn.2018.11.003. Epub 2018 Nov 20. Mol Ther Nucleic Acids. 2019. PMID: 30583098 Free PMC article. - Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre.
Bellini D, Caly DL, McCarthy Y, Bumann M, An SQ, Dow JM, Ryan RP, Walsh MA. Bellini D, et al. Mol Microbiol. 2014 Jan;91(1):26-38. doi: 10.1111/mmi.12447. Epub 2013 Nov 24. Mol Microbiol. 2014. PMID: 24176013 Free PMC article. - cGAS-STING, an important signaling pathway in diseases and their therapy.
Li Q, Wu P, Du Q, Hanif U, Hu H, Li K. Li Q, et al. MedComm (2020). 2024 Mar 23;5(4):e511. doi: 10.1002/mco2.511. eCollection 2024 Apr. MedComm (2020). 2024. PMID: 38525112 Free PMC article. Review.
References
- Keating SE, Baran M, Bowie AG. Trends Immunol. 2011;32:574–81. - PubMed
- Kato H, Takahasi K, Fujita T. Immunol Rev. 2011;243:91–8. - PubMed
- Karaolis DK, et al. J Immunol. 2007;178:2171–81. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials