Perk-dependent repression of miR-106b-25 cluster is required for ER stress-induced apoptosis - PubMed (original) (raw)
doi: 10.1038/cddis.2012.74.
D E Read, A Deepti, K Cawley, A Gupta, D Oommen, T Verfaillie, S Matus, M A Smith, J L Mott, P Agostinis, C Hetz, A Samali
Affiliations
- PMID: 22739985
- PMCID: PMC3388242
- DOI: 10.1038/cddis.2012.74
Perk-dependent repression of miR-106b-25 cluster is required for ER stress-induced apoptosis
S Gupta et al. Cell Death Dis. 2012.
Abstract
Activation of the unfolded protein response sensor PKR-like endoplasmic reticulum kinase (Perk) attenuates endoplasmic reticulum (ER) stress levels. Conversantly, if the damage is too severe and ER function cannot be restored, this signaling branch triggers apoptosis. Bcl-2 homology 3-only family member Bim is essential for ER stress-induced apoptosis. However, the regulatory mechanisms controlling Bim activation under ER stress conditions are not well understood. Here, we show that downregulation of the miR-106b-25 cluster contributes to ER stress-induced apoptosis and the upregulation of Bim. Hypericin-mediated photo-oxidative ER damage induced Perk-dependent cell death and led to a significant decrease in the levels of miRNAs belonging to miR-106b-25 cluster in wild-type (WT) but not in Perk⁻/⁻ MEFs. Further, we show that expression of miR-106b-25 and Mcm-7 (host gene of miR-106b-25) is co-regulated through the transcription factors Atf4 (activating transcription factor 4) and Nrf2 (nuclear factor-erythroid-2-related factor 2). ER stress increased the activity of WT Bim 3'UTR (untranslated region) construct but not the miR-106b-25 recognition site-mutated Bim 3'UTR construct. Overexpression of miR-106b-25 cluster inhibits ER stress-induced cell death in WT but did not confer any further protection in Bim-knockdown cells. Further, we show downregulation in the levels of miR-106b-25 cluster in the symptomatic SOD1(G86R) transgenic mice. Our results suggest a molecular mechanism whereby repression of miR-106b-25 cluster has an important role in ER stress-mediated increase in Bim and apoptosis.
Figures
Figure 1
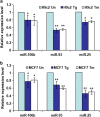
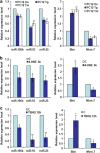
Downregulation of miRNAs of the miR-106b-25 cluster during conditions of ER stress. (a) H9c2 cardiomyocytes were either untreated (Un) or treated with (1.0 _μ_M) Tg and (1.0 _μ_g/ml) Tm for 24 h. The expression level of indicated miRNAs was quantified by real-time RT-PCR, normalizing against snoRNA. Error bars represent mean±S.D. from three independent experiments performed in triplicate. (b) MCF-7 cells were either untreated (Un) or treated with (2.0 _μ_M) Tg and (2.0 _μ_g/ml) Tm for 24 h and the expression level of indicated miRNAs was quantified by real-time RT-PCR, normalizing against U6 snRNA. Error bars represent mean±S.D. from three independent experiments performed in triplicate. (*P<0.05, **P<0.01, two-tailed unpaired _t_-test compared with untreated cells)
Figure 2
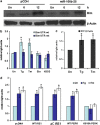
Perk-dependent increases in the activity of Bim 3′UTR reporter construct during conditions of ER stress. (a) pCDH and miR-106b-25 cells were either untreated (Un) or treated with (0.25 _μ_M) Tg for indicated time points. Total protein was isolated and immunoblotting was performed using antibodies against Bim and _β_-actin. (b) MCF-7 cells were transfected with Bim 3′UTR reporter plasmid containing binding sites for members of miR-106b-25 (Bim UTR-wt) or Bim 3′UTR reporter plasmid with mutated binding sites for members of miR-106b-25 (Bim UTR-mt). At 24 h after transfection cells were left untreated (Un) or treated with (2.0 _μ_M) Tg and (2.0 _μ_g/ml) Tm, (50 _μ_g/ml) Eto, or (600 _μ_M) H2O2. Luciferase activity was measured 48 h after transfection using Dual-Glo assay system and normalized luciferase activity (Renilla/Firefly) is shown. Error bars represent mean±S.D. from three independent experiments performed in duplicate. (c) PC12 cells were transfected with Bim 3′UTR reporter plasmid containing binding sites for members of miR-106b-25 (Bim UTR-wt). At 24 h after transfection cells were left untreated (Un) or treated with (0.25 _μ_M) Tg and (2.0 μ_g/ml) Tm. Luciferase activity was measured 48 h after transfection using Dual-Glo assay system and normalized luciferase activity (Renilla/Firefly) is shown. Error bars represent mean±S.D. from three independent experiments performed in duplicate. (d) MCF-7 cells were transfected with Bim 3′UTR reporter plasmid containing binding sites for members of miR-106b-25 (Bim UTR-wt) in combination with pcDNA3 (pcDNA), WT Ire1, dominant-negative Ire1 (Δ_C Ire1), WT Perk or dominant-negative Perk (K618A Perk) expression plasmids. At 24 h after transfection cells were left untreated (Un) or treated with (2.0 _μ_M) Tg and (2.0 _μ_g/ml) Tm. Luciferase activity was measured 48 h after transfection using Dual-Glo assay system and normalized luciferase activity (Renilla/Firefly) is shown. Error bars represent mean±S.D. from three independent experiments performed in duplicate. (*P<0.05, two-tailed unpaired _t_-test compared with untreated cells)
Figure 3
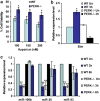
PERK-dependent regulation of miR-106b-25 cluster during conditions of ER stress. (a) WT and Perk knockout (Perk−/−) MEFs were exposed to phOx stress (200 nM hypericin for 2 h, 2.7 J/cm2). The graph shows the surviving fraction of cells 24 h after treatment. Error bars represent mean±S.D. from three experiments performed in duplicate. (*P<0.05, two-tailed unpaired _t_-test compared with WT MEFs). (b) WT and Perk knockout (Perk−/−) MEFs were exposed to phOx stress (200 nM hypericin for 2 h, 2.7J/cm2) and expression levels of Bim was quantified by real-time RT-PCR, normalizing against Gapdh. Error bars represent mean±S.D. from two independent experiments performed in triplicate. (c) WT and Perk knockout (Perk−/−) MEFs were incubated with 200 nM hypericin for 2 h and irradiated (2.7 J/cm2) (phOx stress). Expression level of miR-106b, miR-25, and miR-93 was quantified by real-time RT-PCR, normalizing against snoRNA. Error bars represent mean±S.D. from two independent experiments performed in triplicate. (*P<0.05, **P<0.01, two-tailed unpaired _t_-test compared with untreated WT MEFs)
Figure 4
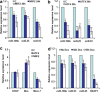
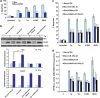
Effect of ectopic Atf4 and Nrf2 on the expression miR-106b-25 cluster and Bim. (a and b) PC12 cells were transfected with pcDNA3 (c), WT Atf4 or Nrf2 expression plasmids and total RNA was isolated at indicated time points. Expression levels of indicated miRNAs were quantified by real-time RT-PCR, normalizing against snoRNA. Error bars represent mean±S.D. from three independent experiments performed in triplicate. (*P<0.05, **P<0.01, two-tailed unpaired _t_-test as compared with control transfected cells). (c) PC12 cells were transfected with pcDNA3 (c), WT Atf4, or Nrf2 expression plasmids. Total RNA was isolated and expression levels of Chop, Bim, and Mcm-7 were quantified by real-time RT-PCR, normalizing against Gapdh. Error bars represent mean±S.D. from three independent experiments performed in triplicate. (*P<0.05, **P<0.01, two-tailed unpaired _t_-test as compared with control transfected cells). (d) pTRIPZ-shKeap1-293T cells were treated with (500 ng/ml) of doxycycline for indicated time points and expression levels of miR-106b, miR-93, and miR-25 were quantified by real-time RT-PCR, normalizing against snoRNA. Error bars represent mean±S.D. from four independent experiments performed in triplicate. (*P<0.05, **P<0.01, two-tailed unpaired _t_-test as compared with uninduced (No Dox) cells)
Figure 5
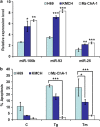
Downregulation of miRNAs belonging to miR-106b-25 cluster during conditions of ER stress. PC12 cells were either untreated (Un) or treated with (0.25 _μ_M) Tg and (2.0 _μ_g/ml) Tm for 24 h (a), (100 _μ_M) 4-HNE for 6 h (b), and (0.5 mM) tBHQ for 12 h (c). The expression levels of miR-106b, miR-93 and miR-25 were quantified by real-time RT-PCR, normalizing against snoRNA. The expression level of Bim and Mcm-7 was quantified by real-time RT-PCR, normalizing against Gapdh. Error bars represent mean±S.D. from three independent experiments performed in triplicate. (*P<0.05, **P<0.01, two-tailed unpaired _t_-test as compared with untreated cells)
Figure 6
Inhibition of ER stress-induced apoptosis by miR-106b-25 cluster is mediated by repression of Bim. (a) The control (Neo) and Bim-shRNA expressing (Bim-shRNA) PC12 cells were either untreated (Un) or treated with (0.25 _μ_M) Tg, (2.0 _μ_g/ml) Tm, (0.5 mM) tBHQ, and (100 _μ_M) 4-4-HNE for 48 h. Apoptosis was determined with annexin-V/PI staining followed by FACS analysis. Percentages of cells positive for both annexin-V and annexin-V/PI are shown. Average and error bars represent mean±S.D. from three independent experiments. (*P<0.005, two-tailed unpaired _t_-test as compared with control (Neo) cells). (b) Upper panel, The indicated cells were either untreated (Un) or treated with (0.25 _μ_M) Tg for indicated 24 h. Total protein was isolated and immunoblotting was performed using antibodies against Bim, Chop and _β_-actin. Lower panel, in the experiment described in (a), induction of Bim and Chop was calculated after densitometric analysis of the autoradiographs with Image J. Average and error bars represent mean±S.D. from three independent experiments.(**P<0.05, two-tailed unpaired _t_-test as compared with miR-106b-25 expressing (Neo/miR-106b-25) cells). (c) The indicated cells were either untreated or treated with (0.25 _μ_M) Tg, (2.0 _μ_g/ml) Tm, (0.5 mM) tBHQ, and (100 _μ_M) 4-4-HNE for 48 h. Apoptosis was determined with annexin-V/PI staining followed by FACS analysis. Percentages of cells positive for both annexin-V and annexin-V/PI are shown. Average and error bars represent mean±S.D. from three independent experiments. (*P<0.005, two-tailed unpaired _t_-test as compared with control (Neo/pCDH) cells). (d) The indicated cells were either untreated or treated with (0.25 _μ_M) Tg for 24 h or (2.0 _μ_g/ml) Tm for 24 h, (0.5 mM) tBHQ for 12 h, and (100 _μ_M) 4-4-HNE for 12 h, and DEVDase activity was measured as described in Materials and Methods. Average and error bars represent mean±S.D. from three independent experiments. (*P<0.005, two-tailed unpaired _t_-test as compared with control (Neo/pCDH) cells)
Figure 7
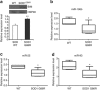
Downregulation of miRNAs belonging to miR-106b-25 cluster in the SOD1 mutant mice. (a) The total protein was isolated from the cortex of WT and symptomatic SOD1G86R mice. A total of 40 _μ_g protein per sample used for immunoblotting was analyzed using antibodies against Bim and HSP90. Upper panel, western blot analysis of Bim expression in cortex of WT and symptomatic SOD1G86R mice. Lower panel, average of Bim expression in the cortex of WT (_n_=2) and symptomatic SOD1G86R (_n_=7) mice are shown and error bars represent mean±S.D. (**P<0.05, two-tailed unpaired _t_-test as compared with WT mice). (b–d) The total RNA was isolated from the cortex of WT and symptomatic SOD1G86R mice and expression levels of miR-106b, miR-93, and miR-25 were quantified by real-time RT-PCR, normalizing against U6 snRNA. Box plots showing median and distribution of miR-106b (b), miR-25, (c) and miR-93 (d) in the cortex of WT (_n_=6) and symptomatic SOD1G86R (_n_=6) mice are shown. (*P<0.05, Wilcoxon signed rank test as compared with WT mice)
Figure 8
Effect of miR-106b-25 expression on sensitivity to ER stress-induced cell death. (a) The expression of miR-106b-25 cluster members was measured by qRT-PCR from total RNA isolated from lysates of the indicated cell lines. Results are presented as relative expression using Z30 as an internal control and employing the delta-delta _C_t method. Error bars represent mean±S.E. from three independent experiments performed in triplicate. Statistical significance was determined by ANOVA with Bonferroni post hoc correction; *P<0.05, **P<0.01, ***P<0.001, compared with H69 cells. (b) Indicated cell lines were treated with (2.0 _μ_M) Tg and (2.0 _μ_g/ml) Tm for 24 h. After treatment the apoptotic nuclei were determined as described in Materials and Methods section. Error bars represent mean±S.E. from three independent experiments performed in triplicate. Statistical significance was determined by ANOVA with Bonferroni post hoc correction; *P<0.05, **P<0.01, ***P<0.001, compared with H69 cells
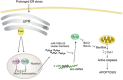
Figure 9
Schematic summary detailing the main conclusions of this manuscript. During ER stress the transcription factors Atf4 and Nrf2 are activated and downregulate the Mcm7, host gene of miR-106b-25 cluster in a Perk dependent manner, thus contributing to induction of Bim and ER stress-induced apoptosis (see text for details)
Similar articles
- TSA suppresses miR-106b-93-25 cluster expression through downregulation of MYC and inhibits proliferation and induces apoptosis in human EMC.
Zhao ZN, Bai JX, Zhou Q, Yan B, Qin WW, Jia LT, Meng YL, Jin BQ, Yao LB, Wang T, Yang AG. Zhao ZN, et al. PLoS One. 2012;7(9):e45133. doi: 10.1371/journal.pone.0045133. Epub 2012 Sep 19. PLoS One. 2012. PMID: 23028803 Free PMC article. - PERK-mediated induction of microRNA-483 disrupts cellular ATP homeostasis during the unfolded protein response.
Hiramatsu N, Chiang K, Aivati C, Rodvold JJ, Lee JM, Han J, Chea L, Zanetti M, Koo EH, Lin JH. Hiramatsu N, et al. J Biol Chem. 2020 Jan 3;295(1):237-249. doi: 10.1074/jbc.RA119.008336. Epub 2019 Dec 2. J Biol Chem. 2020. PMID: 31792031 Free PMC article. - Endoplasmic reticulum stress signaling is involved in mitomycin C (MMC)-induced apoptosis in human fibroblasts via PERK pathway.
Shi K, Wang D, Cao X, Ge Y. Shi K, et al. PLoS One. 2013;8(3):e59330. doi: 10.1371/journal.pone.0059330. Epub 2013 Mar 22. PLoS One. 2013. PMID: 23533616 Free PMC article. - The Role of the PERK/eIF2α/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress.
Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, Majsterek I. Rozpedek W, et al. Curr Mol Med. 2016;16(6):533-44. doi: 10.2174/1566524016666160523143937. Curr Mol Med. 2016. PMID: 27211800 Free PMC article. Review. - Stress relief for cancer immunotherapy: implications for the ER stress response in tumor immunity.
Andrews AM, Tennant MD, Thaxton JE. Andrews AM, et al. Cancer Immunol Immunother. 2021 May;70(5):1165-1175. doi: 10.1007/s00262-020-02740-3. Epub 2020 Oct 26. Cancer Immunol Immunother. 2021. PMID: 33104836 Free PMC article. Review.
Cited by
- Inherited cataracts: Genetic mechanisms and pathways new and old.
Shiels A, Hejtmancik JF. Shiels A, et al. Exp Eye Res. 2021 Aug;209:108662. doi: 10.1016/j.exer.2021.108662. Epub 2021 Jun 12. Exp Eye Res. 2021. PMID: 34126080 Free PMC article. Review. - UPR-Induced miR-616 Inhibits Human Breast Cancer Cell Growth and Migration by Targeting c-MYC.
Arabkari V, Sultana A, Barua D, Webber M, Smith T, Gupta A, Gupta S. Arabkari V, et al. Int J Mol Sci. 2023 Aug 22;24(17):13034. doi: 10.3390/ijms241713034. Int J Mol Sci. 2023. PMID: 37685841 Free PMC article. - miR-106b-responsive gene landscape identifies regulation of Kruppel-like factor family.
Wehrkamp CJ, Natarajan SK, Mohr AM, Phillippi MA, Mott JL. Wehrkamp CJ, et al. RNA Biol. 2018 Mar 4;15(3):391-403. doi: 10.1080/15476286.2017.1422471. Epub 2018 Feb 1. RNA Biol. 2018. PMID: 29286255 Free PMC article. - Mir106b-25 and Mir17-92 Are Crucially Involved in the Development of Experimental Neuroinflammation.
Finardi A, Diceglie M, Carbone L, Arnò C, Mandelli A, De Santis G, Fedeli M, Dellabona P, Casorati G, Furlan R. Finardi A, et al. Front Neurol. 2020 Aug 21;11:912. doi: 10.3389/fneur.2020.00912. eCollection 2020. Front Neurol. 2020. PMID: 32973667 Free PMC article. - A blood miRNA signature associates with sporadic Creutzfeldt-Jakob disease diagnosis.
Norsworthy PJ, Thompson AGB, Mok TH, Guntoro F, Dabin LC, Nihat A, Paterson RW, Schott JM, Collinge J, Mead S, Viré EA. Norsworthy PJ, et al. Nat Commun. 2020 Aug 7;11(1):3960. doi: 10.1038/s41467-020-17655-x. Nat Commun. 2020. PMID: 32769986 Free PMC article.
References
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. - PubMed
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. - PubMed
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. - PubMed
- Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–1349. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous