Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion - PubMed (original) (raw)
Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion
Eduardo Cebollero et al. Curr Biol. 2012.
Abstract
Background: The biogenesis of autophagosomes, the hallmark of autophagy, depends on the function of the autophagy-related (Atg) proteins and the generation of phosphatidylinositol-3-phosphate (PtdIns3P) at the phagophore assembly site (PAS), the location where autophagosomes arise. The current model is that PtdIns3P is involved primarily in the recruitment of Atg proteins to the PAS and that once an autophagosome is complete, the Atg machinery is released from its surface back into the cytoplasm and reused for the formation of new vesicles.
Results: We have identified a PtdIns3P phosphatase, Ymr1, that is essential for the normal progression of both bulk and selective types of autophagy. This protein is recruited to the PAS at an early stage of formation of this structure through a process that requires both its GRAM domain and its catalytic activity. In the absence of Ymr1, Atg proteins fail to dissociate from the limiting membrane of autophagosomes, and these vesicles accumulate in the cytoplasm.
Conclusions: Our data thus reveal a key role for PtdIns3P turnover in the regulation of the late steps of autophagosome biogenesis and indicate that the disassembly of the Atg machinery from the surface of autophagosomes is a requisite for their fusion with the vacuole.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
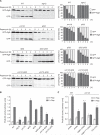
Figure 1. PtdIns3P Phosphatases Are Essential for Autophagy
(A) WT (SEY6210), atg1Δ (WHY1), ymr1Δ (YMR1Δ), sjl2Δ (JGY130), sjl3Δ (JGY131), sjl2Δ sjl3Δ (JGY132), ymr1Δ sjl2Δ (BPY06), and ymr1Δ sjl3Δ (BPY01) strains carrying the pCuGFPAtg8(416) plasmid were incubated with rapamycin (Rap) to induce autophagy and culture aliquots were collected at intervals of 1 hr. GFP-Atg8 cleavage was determined by western blot with anti-GFP antibodies, and the percentages of GFP-Atg8 and GFP were plotted. (B) WT (YTS159), atg9Δ (FRY300), ymr1Δ (ECY190), sjl2Δ (ECY194), sjl3Δ (ECY189), sjl2Δ sjl3Δ (ECY192), ymr1Δ sjl2Δ (ECY200), and ymr1Δ sjl3Δ (ECY193) cells expressing Pho8D60 were grown and treated as in (A) before measuring Pho8Δ60 activity. (C) The WT (YTS159) or the ymr1Δ (ECY190) strain harboring pRS426 (empty), pYMR1(416) [YMR1(CEN)], pYMR1MUT(416) [YMR1C397S(CEN)], or pRSSJL3(426) [SJL3(2μ)] plasmids were processed as in (B). The graphs represent the average of three experiments ± SEM. The asterisks indicate a significant difference with the WT, while the number sign shows a significant difference with the ymr1Δ strain harboring pRS426 (p values < 0.05). See also Figure S1.
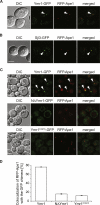
Figure 2. The PH-GRAM Domain and Phosphatase Activity Are Required for Ymr1 Localization to the PAS
(A) WT cells expressing RFP-Ape1 and Ymr1-GFP (ECY202) were incubated with rapamycin for 3 hr before collecting images. (B) Cells expressing RFP-Ape1 and Sjl3-GFP (AVY070) were treated and imaged as in (A). (C) The ymr1Δ RFP-Ape1 (ECY207) strain carrying integrated pYMR1GFP(406) (Ymr1-GFP), pNΔYMR1GFP(406) (NΔYmr1-GFP), or pYMR1C397SGFP(406) (Ymr1C397S-GFP) were starved for 3 hr in SD-N medium before imaging. Arrows highlight colocalization. DIC, differential interference contrast. (D) Quantification of the degree of RFP-Ape1 colocalization with the GFP-tagged proteins in the samples imaged in (C). The graphs represent the average of two experiments ± SEM. Scale bars represent 5 μm. See also Figure S2.
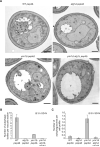
Figure 3. Electron Microscopy Analysis of the ymr1Δ Strain
WT pep4Δ (TVY1), atg1Δ pep4Δ (YTS113), ymr1Δ pep4Δ (AVY059), and ymr1Δ atg1Δ pep4Δ (ECY191) cells were grown in rich medium to early log phase and then transferred into SD-N medium for 3 hr before processing the samples for EM as described in the Experimental Procedures. (A) Micrographs of starved cells. Autophagic bodies are marked with an asterisk, while an autophagosome is highlighted with an arrowhead. N, nucleus; V, vacuole. Scale bars represent 500 nm. (B) Quantification of the autophagic bodies. (C) Quantification of autophagosome accumulation. In (B) and (C), the results are expressed as the average number of autophagic bodies per vacuole and autophagosomes per cell section, respectively. Error bars represent the SD in the counting of two different grids. See also Figure S3.
Figure 4. Closed Autophagosomes Accumulate in the Absence of Ymr1
(A) Schematic explaining the principle of the prApe1 protease protection assay. (B) The WT (SEY6210), vam3Δ (CWY40), atg1Δ (WHY1), and ymr1Δ (YMR1Δ) cells were converted to spheroplasts and starved for 3 hr in SD-N. Total cell extracts from lysed spheroplasts were centrifuged and the pellet fraction was either not treated, or mixed with proteinase K (PK) and/or TX-100 detergent before being incubated on ice for 30 min. After protein precipitation, samples were analyzed by immunoblots with anti-prApe1 antibodies [38]. The experiment was repeated four times with identical results.
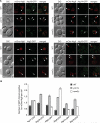
Figure 5. Atg Proteins Remain Associated with Autophagosomes in the ymr1Δ Strain
WT, ymr1Δ, and vam3Δ strains expressing endogenous Atg2-GFP (PSY102, ECY155, and ECY183), Atg9-GFP (FRY162, ECY153, and AVY078) Atg14-GFP (PSY142, ECY162, and ECY184), Atg16-GFP (KTY148, ECY157, and ECY185), Atg17-GFP (ECY167, ECY169, and ECY172), or Atg18-GFP (PSY62, ECY137, and ECY147) and the pCumCheAtg8(415) plasmid were treated with rapamycin for 3 hr and imaged. (A) Fluorescence microscopy images of the various strains. Arrows highlight colocalizations. DIC, differential interference contrast. Scale bars represent 5 μm. (B) Quantification of the experiments presented in (A) and Figure S4A by determining the average number of GFP chimera positive for mCheAtg8 per cell. Error bars represent the SEM. See also Figure S4.
Figure 6. Atg8 Dynamic Association with the PAS Is Impaired in the Absence of YMR1
(A) PtdIns3P turnover is important for Atg8 dissociation from the PAS. WT and ymr1Δ cells expressing endogenous GFP-Atg8 (MZY089 and MZY090) were exposed for 3 hr to rapamycin before being analyzed by microscopy. The cycle of GFP-Atg8 at the PAS was determined by quantification of the time elapsed from the appearance of a GFP-Atg8 punctum until it disappears. All data points came from four independent experiments; error bars indicate the SEM, and asterisks show significant differences. (B) Atg8 accumulates at the PAS in the absence of YMR1. The fluorescence intensity was calculated as described in the Experimental Procedures. Data points came from three independent experiments and error bars indicate the SEM. (C) Proposed model describing the role of Ymr1 during autophagosome biogenesis. This phosphatase is recruited at the early stage of PAS formation, when all the Atg proteins start to assemble following a hierarchical order at this site [7]. The Atg machinery first mediates the formation of the phagophore and then expands it into an immature autophagosome. The release of the Atg proteins and Ymr1, a step that is at least in part regulated through the hydrolysis of PtdIns3P into PtdIns by Ymr1 and probably required for the recycling of the Atg machinery, leads to the completion of the autophagosome, which is then ready to fuse with the vacuole. Our data cannot exclude a function of Ymr1 during the initial phases of the Atg machinery assembly at the PAS (question mark).
Similar articles
- PI3P phosphatase activity is required for autophagosome maturation and autolysosome formation.
Wu Y, Cheng S, Zhao H, Zou W, Yoshina S, Mitani S, Zhang H, Wang X. Wu Y, et al. EMBO Rep. 2014 Sep;15(9):973-81. doi: 10.15252/embr.201438618. Epub 2014 Aug 14. EMBO Rep. 2014. PMID: 25124690 Free PMC article. - Understanding phosphatidylinositol-3-phosphate dynamics during autophagosome biogenesis.
Cebollero E, van der Vaart A, Reggiori F. Cebollero E, et al. Autophagy. 2012 Dec;8(12):1868-70. doi: 10.4161/auto.22162. Epub 2012 Sep 19. Autophagy. 2012. PMID: 22992453 Free PMC article. - WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome.
Proikas-Cezanne T, Takacs Z, Dönnes P, Kohlbacher O. Proikas-Cezanne T, et al. J Cell Sci. 2015 Jan 15;128(2):207-17. doi: 10.1242/jcs.146258. J Cell Sci. 2015. PMID: 25568150 Review. - Local detection of PtdIns3P at autophagosome biogenesis membrane platforms.
Nascimbeni AC, Codogno P, Morel E. Nascimbeni AC, et al. Autophagy. 2017 Sep 2;13(9):1602-1612. doi: 10.1080/15548627.2017.1341465. Epub 2017 Aug 16. Autophagy. 2017. PMID: 28813193 Free PMC article. - Mechanisms of autophagy and pexophagy in yeasts.
Sibirny AA. Sibirny AA. Biochemistry (Mosc). 2011 Dec;76(12):1279-90. doi: 10.1134/S0006297911120017. Biochemistry (Mosc). 2011. PMID: 22150273 Review.
Cited by
- The yeast Saccharomyces cerevisiae: an overview of methods to study autophagy progression.
Delorme-Axford E, Guimaraes RS, Reggiori F, Klionsky DJ. Delorme-Axford E, et al. Methods. 2015 Mar;75:3-12. doi: 10.1016/j.ymeth.2014.12.008. Epub 2014 Dec 16. Methods. 2015. PMID: 25526918 Free PMC article. Review. - Atg1-dependent phosphorylation of Vps34 is required for dynamic regulation of the phagophore assembly site and autophagy in Saccharomyces cerevisiae.
Lee Y, Kim B, Jang HS, Huh WK. Lee Y, et al. Autophagy. 2023 Sep;19(9):2428-2442. doi: 10.1080/15548627.2023.2182478. Epub 2023 Mar 2. Autophagy. 2023. PMID: 36803233 Free PMC article. - Autophagosomes are formed at a distinct cellular structure.
Hollenstein DM, Kraft C. Hollenstein DM, et al. Curr Opin Cell Biol. 2020 Aug;65:50-57. doi: 10.1016/j.ceb.2020.02.012. Epub 2020 Mar 20. Curr Opin Cell Biol. 2020. PMID: 32203894 Free PMC article. Review. - Posttranslational modification of autophagy-related proteins in macroautophagy.
Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, Tang D. Xie Y, et al. Autophagy. 2015;11(1):28-45. doi: 10.4161/15548627.2014.984267. Autophagy. 2015. PMID: 25484070 Free PMC article. Review. - PI3P phosphatase activity is required for autophagosome maturation and autolysosome formation.
Wu Y, Cheng S, Zhao H, Zou W, Yoshina S, Mitani S, Zhang H, Wang X. Wu Y, et al. EMBO Rep. 2014 Sep;15(9):973-81. doi: 10.15252/embr.201438618. Epub 2014 Aug 14. EMBO Rep. 2014. PMID: 25124690 Free PMC article.
References
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009;10:458–467. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources