RNA degradation in Saccharomyces cerevisae - PubMed (original) (raw)
Review
RNA degradation in Saccharomyces cerevisae
Roy Parker. Genetics. 2012 Jul.
Abstract
All RNA species in yeast cells are subject to turnover. Work over the past 20 years has defined degradation mechanisms for messenger RNAs, transfer RNAs, ribosomal RNAs, and noncoding RNAs. In addition, numerous quality control mechanisms that target aberrant RNAs have been identified. Generally, each decay mechanism contains factors that funnel RNA substrates to abundant exo- and/or endonucleases. Key issues for future work include determining the mechanisms that control the specificity of RNA degradation and how RNA degradation processes interact with translation, RNA transport, and other cellular processes.
Figures
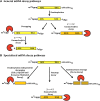
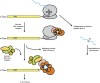
Figure 1
(A) General mRNA decay pathways. (B) Specialized mRNA decay pathways.
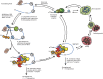
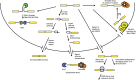
Figure 2
Model for mRNA decapping.
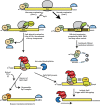
Figure 3
Model for the nonsense-mediated decay.
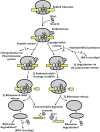
Figure 4
Model for no-go decay.
Figure 5
Model for non-stop decay.
Figure 6
Mechanisms of degradation for unspliced pre-mRNAs.
Similar articles
- Sequential RNA degradation pathways provide a fail-safe mechanism to limit the accumulation of unspliced transcripts in Saccharomyces cerevisiae.
Sayani S, Chanfreau GF. Sayani S, et al. RNA. 2012 Aug;18(8):1563-72. doi: 10.1261/rna.033779.112. Epub 2012 Jul 2. RNA. 2012. PMID: 22753783 Free PMC article. - Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution.
Maderazo AB, Belk JP, He F, Jacobson A. Maderazo AB, et al. Mol Cell Biol. 2003 Feb;23(3):842-51. doi: 10.1128/MCB.23.3.842-851.2003. Mol Cell Biol. 2003. PMID: 12529390 Free PMC article. - In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation.
Aw JG, Shen Y, Wilm A, Sun M, Lim XN, Boon KL, Tapsin S, Chan YS, Tan CP, Sim AY, Zhang T, Susanto TT, Fu Z, Nagarajan N, Wan Y. Aw JG, et al. Mol Cell. 2016 May 19;62(4):603-17. doi: 10.1016/j.molcel.2016.04.028. Epub 2016 May 12. Mol Cell. 2016. PMID: 27184079 - Molecular chaperones and quality control in noncoding RNA biogenesis.
Wolin SL, Wurtmann EJ. Wolin SL, et al. Cold Spring Harb Symp Quant Biol. 2006;71:505-11. doi: 10.1101/sqb.2006.71.051. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381333 Review. - Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae.
Tucker M, Parker R. Tucker M, et al. Annu Rev Biochem. 2000;69:571-95. doi: 10.1146/annurev.biochem.69.1.571. Annu Rev Biochem. 2000. PMID: 10966469 Review.
Cited by
- Quality control of chemically damaged RNA.
Simms CL, Zaher HS. Simms CL, et al. Cell Mol Life Sci. 2016 Oct;73(19):3639-53. doi: 10.1007/s00018-016-2261-7. Epub 2016 May 7. Cell Mol Life Sci. 2016. PMID: 27155660 Free PMC article. Review. - Widespread Co-translational RNA Decay Reveals Ribosome Dynamics.
Pelechano V, Wei W, Steinmetz LM. Pelechano V, et al. Cell. 2015 Jun 4;161(6):1400-12. doi: 10.1016/j.cell.2015.05.008. Cell. 2015. PMID: 26046441 Free PMC article. - Pbp1 is involved in Ccr4- and Khd1-mediated regulation of cell growth through association with ribosomal proteins Rpl12a and Rpl12b.
Kimura Y, Irie K, Irie K. Kimura Y, et al. Eukaryot Cell. 2013 Jun;12(6):864-74. doi: 10.1128/EC.00370-12. Epub 2013 Apr 5. Eukaryot Cell. 2013. PMID: 23563484 Free PMC article. - Compositional Control of Phase-Separated Cellular Bodies.
Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK. Banani SF, et al. Cell. 2016 Jul 28;166(3):651-663. doi: 10.1016/j.cell.2016.06.010. Epub 2016 Jun 30. Cell. 2016. PMID: 27374333 Free PMC article. - Ccr4-not regulates RNA polymerase I transcription and couples nutrient signaling to the control of ribosomal RNA biogenesis.
Laribee RN, Hosni-Ahmed A, Workman JJ, Chen H. Laribee RN, et al. PLoS Genet. 2015 Mar 27;11(3):e1005113. doi: 10.1371/journal.pgen.1005113. eCollection 2015 Mar. PLoS Genet. 2015. PMID: 25815716 Free PMC article.
References
- Aebi M., Kirchner G., Chen J. Y., Vijayraghavan U., Jacobson A., et al. , 1990. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 265: 16216–16220 - PubMed
- Alexandrov A., Chernyakov I., Gu W., Hiley S. L., Hughes T. R., et al. , 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 21: 87–96 - PubMed
- Amrani N., Ganesan R., Kervestin S., Mangus D. A., Ghosh S., et al. , 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous