Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues - PubMed (original) (raw)
Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues
Sebastian A Wagner et al. Mol Cell Proteomics. 2012 Dec.
Abstract
Posttranslational modifications of proteins increase the complexity of the cellular proteome and enable rapid regulation of protein functions in response to environmental changes. Protein ubiquitylation is a central regulatory posttranslational modification that controls numerous biological processes including proteasomal degradation of proteins, DNA damage repair and innate immune responses. Here we combine high-resolution mass spectrometry with single-step immunoenrichment of di-glycine modified peptides for mapping of endogenous putative ubiquitylation sites in murine tissues. We identify more than 20,000 unique ubiquitylation sites on proteins involved in diverse biological processes. Our data reveals that ubiquitylation regulates core signaling pathways common for each of the studied tissues. In addition, we discover that ubiquitylation regulates tissue-specific signaling networks. Many tissue-specific ubiquitylation sites were obtained from brain highlighting the complexity and unique physiology of this organ. We further demonstrate that different di-glycine-lysine-specific monoclonal antibodies exhibit sequence preferences, and that their complementary use increases the depth of ubiquitylation site analysis, thereby providing a more unbiased view of protein ubiquitylation.
Figures
Fig. 1.
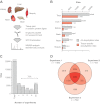
Identification of endogenous ubiquitylation sites in murine tissues by mass spectrometry. A, Frozen tissues were cryogrinded and proteins were solubilized in modified radioimmunoprecipitation assay buffer. Proteins were digested using trypsin, and di-glycine modified peptides were enriched using two different di-glycine-lysine-specific monoclonal antibodies. Enriched peptides were fractionated using strong-cation exchange chromatography and analyzed by mass spectrometry. B, The bar chart illustrates the total number of ubiquitylation sites and the number of tissue-specific ubiquitylation sites identified in each tissue. The number of ubiquitylation sites identified from all tissues (except liver) is combined from two independent replicate experiments. The number of ubiquitylation sites identified from liver tissue is combined from three independent replicate experiments (indicated by an asterisk). C, Number of experiments in which individual sites were found. Over 70% of the reported sites were found in two or more experiments. D, Venn diagram illustrating the experimental overlap between the three replicate experiments performed from liver tissue.
Fig. 2.
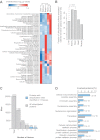
Ubiquitylated proteins participate in tissue-specific biological processes. A, The enrichment of GO biological process terms among ubiquitylated proteins in different tissues was analyzed and illustrated as heat map. Only sites that were found in two or more replicate experiments were used for the analysis. The complete results of the analysis can be found in
supplemental Table S2
. B, The bar chart illustrates the fraction of ubiquitylation sites on plasma membrane proteins in different tissues. All proteins annotated with the GO cellular compartment term “plasma membrane” were considered as plasma membrane proteins. Fisher exact test was applied to determine the significance of the observed differences. C, The bar chart illustrates the number of ubiquitylation sites detected in one, two, three, four, or five tissues. Sites identified in at least four out of five tissues are indicated with light blue bars. D, GO biological process term enrichment analysis of proteins with ubiquitylation sites that were identified in at least four of the studied tissues.
Fig. 3.
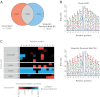
Sequence preferences of di-glycine-specific monoclonal antibodies. A, Number of ubiquitylation sites identified with the two different antibodies. The red and blue area illustrates the ubiquitylation sites identified only with the monoclonal antibody clone GX41 or the Ubiquitin Remnant Motif Kit. B, Sequence motif analysis of ubiquitylation sites identified exclusively with the monoclonal antibody clone GX41 or the Ubiquitin Remnant Motif Kit, respectively. C, Amino acid preferences exhibited by the different di-glycine-lysine antibodies. The relative frequencies of amino acids flanking ubiquitylation sites identified with the two different antibodies were compared using IceLogo. Amino acids that were found overrepresented near di-glycine-lysines identified with the monoclonal antibody clone GX41 are colored in red. Amino acids overrepresented in proximity to ubiquitylation sites identified with the Ubiquitin Remnant Motif Kit are shown in blue. Only amino acids with significant overrepresentation are shown.
Similar articles
- A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles.
Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. Wagner SA, et al. Mol Cell Proteomics. 2011 Oct;10(10):M111.013284. doi: 10.1074/mcp.M111.013284. Epub 2011 Sep 1. Mol Cell Proteomics. 2011. PMID: 21890473 Free PMC article. - Improvement of ubiquitylation site detection by Orbitrap mass spectrometry.
van der Wal L, Bezstarosti K, Sap KA, Dekkers DHW, Rijkers E, Mientjes E, Elgersma Y, Demmers JAA. van der Wal L, et al. J Proteomics. 2018 Feb 10;172:49-56. doi: 10.1016/j.jprot.2017.10.014. Epub 2017 Nov 6. J Proteomics. 2018. PMID: 29122726 - Proteome-wide identification of ubiquitylation sites by conjugation of engineered lysine-less ubiquitin.
Oshikawa K, Matsumoto M, Oyamada K, Nakayama KI. Oshikawa K, et al. J Proteome Res. 2012 Feb 3;11(2):796-807. doi: 10.1021/pr200668y. Epub 2011 Nov 23. J Proteome Res. 2012. PMID: 22053931 - Advances in characterizing ubiquitylation sites by mass spectrometry.
Sylvestersen KB, Young C, Nielsen ML. Sylvestersen KB, et al. Curr Opin Chem Biol. 2013 Feb;17(1):49-58. doi: 10.1016/j.cbpa.2012.12.009. Epub 2013 Jan 5. Curr Opin Chem Biol. 2013. PMID: 23298953 Review. - Ubiquitylomics: An Emerging Approach for Profiling Protein Ubiquitylation in Skeletal Muscle.
Lord SO, Johnston HE, Samant RS, Lai YC. Lord SO, et al. J Cachexia Sarcopenia Muscle. 2024 Dec;15(6):2281-2294. doi: 10.1002/jcsm.13601. Epub 2024 Sep 16. J Cachexia Sarcopenia Muscle. 2024. PMID: 39279720 Free PMC article. Review.
Cited by
- Characterization of the structural determinants of the ubiquitin-dependent proteasomal degradation of human hepatic tryptophan 2,3-dioxygenase.
Liu Y, Kim SM, Wang Y, Karkashon S, Lewis-Ballester A, Yeh SR, Correia MA. Liu Y, et al. Biochem J. 2021 May 28;478(10):1999-2017. doi: 10.1042/BCJ20210213. Biochem J. 2021. PMID: 33960368 Free PMC article. - Role of the alternative splice variant of NCC in blood pressure control.
Wardak H, Tutakhel OAZ, Van Der Wijst J. Wardak H, et al. Channels (Austin). 2018;12(1):346-355. doi: 10.1080/19336950.2018.1528820. Channels (Austin). 2018. PMID: 30264650 Free PMC article. Review. - Cellular Processing of the ABCG2 Transporter-Potential Effects on Gout and Drug Metabolism.
Mózner O, Bartos Z, Zámbó B, Homolya L, Hegedűs T, Sarkadi B. Mózner O, et al. Cells. 2019 Oct 8;8(10):1215. doi: 10.3390/cells8101215. Cells. 2019. PMID: 31597297 Free PMC article. Review. - Identification of chloride intracellular channels as prognostic factors correlated with immune infiltration in hepatocellular carcinoma using bioinformatics analysis.
Huang JJ, Lin J, Chen X, Zhu W. Huang JJ, et al. Medicine (Baltimore). 2021 Nov 12;100(45):e27739. doi: 10.1097/MD.0000000000027739. Medicine (Baltimore). 2021. PMID: 34766585 Free PMC article. - Regulation of cardiac proteasomes by ubiquitination, SUMOylation, and beyond.
Cui Z, Scruggs SB, Gilda JE, Ping P, Gomes AV. Cui Z, et al. J Mol Cell Cardiol. 2014 Jun;71:32-42. doi: 10.1016/j.yjmcc.2013.10.008. Epub 2013 Oct 17. J Mol Cell Cardiol. 2014. PMID: 24140722 Free PMC article. Review.
References
- Pickart C. M., Eddins M. J. (2004) Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 - PubMed
- Hochstrasser M. (1995) Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr. Opin. Cell Biol. 7, 215–223 - PubMed
- Coux O., Tanaka K., Goldberg A. L. (1996) Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65, 801–847 - PubMed
- Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 - PubMed
- Bergink S., Jentsch S. (2009) Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458, 461–467 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases