Automatic annotation of histopathological images using a latent topic model based on non-negative matrix factorization - PubMed (original) (raw)
Automatic annotation of histopathological images using a latent topic model based on non-negative matrix factorization
Angel Cruz-Roa et al. J Pathol Inform. 2011.
Abstract
Histopathological images are an important resource for clinical diagnosis and biomedical research. From an image understanding point of view, the automatic annotation of these images is a challenging problem. This paper presents a new method for automatic histopathological image annotation based on three complementary strategies, first, a part-based image representation, called the bag of features, which takes advantage of the natural redundancy of histopathological images for capturing the fundamental patterns of biological structures, second, a latent topic model, based on non-negative matrix factorization, which captures the high-level visual patterns hidden in the image, and, third, a probabilistic annotation model that links visual appearance of morphological and architectural features associated to 10 histopathological image annotations. The method was evaluated using 1,604 annotated images of skin tissues, which included normal and pathological architectural and morphological features, obtaining a recall of 74% and a precision of 50%, which improved a baseline annotation method based on support vector machines in a 64% and 24%, respectively.
Keywords: Automatic Annotation; Bag of Features; Basal Cell Carcinoma; Histopathology Images; Non-negative Matrix Factorization; Visual Latent Semantic Analysis.
Figures
Figure 1
Example of histopathological images globally annotated with multiple annotations (multilabeled images). These images correspond to the test data set used in this work and they have a resolution of 1024 × 768 pixels. Histopathological annotations of morphological and architectural features such as epidermis, collagen, and hair follicles appear in different images illustrating the high-visual variability for the same annotation
Figure 2
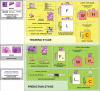
Overview of the proposed method for automatic annotation of histopathological images based on non-negative matrix factorization
Figure 3
Bag of feature setup used for representing histopathology images. In this work the local features extraction is performed using regular grid extraction and each patch of 8 × 8 pixels is represented by the first coefficients of a discrete cosine transform applied to each color component (RGB) independently, the visual codebook is built using k-means with k = 700, and finally each image is represented by a histogram of 700 bins normalized with L1 norm
Figure 4
Examples of training images with the corresponding histopathological annotations. These images have a resolution of 300 × 300 pixels and exhibit only one annotation per image
Figure 5
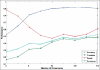
Performance evaluation on training mono-label images by each number of dimensions in the latent space
Figure 6
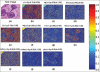
Example of an image from the test data set automatically annotated by the proposed method. The original multilabel image (a) is showed with the salient maps of the patches inside the image according with each one of the 10 histopathological annotations: collagen (b), sebaceous glands (c), hair follicles (d), inflammatory infiltration (e), eccrine glands (f), epidermis (g), nodular basal cell carcinoma (h), morpheiform basal cell carcinoma (i), micro-nodular basal cell carcinoma (j), cystic basal cell carcinoma (k), on the top of each salient image is the real membership of the class (v), the conditional probability estimated by the proposed method (p), and the final concept binarization value (b)
Similar articles
- Multilabel image annotation based on double-layer PLSA model.
Zhang J, Li D, Hu W, Chen Z, Yuan Y. Zhang J, et al. ScientificWorldJournal. 2014;2014:494387. doi: 10.1155/2014/494387. Epub 2014 Jun 4. ScientificWorldJournal. 2014. PMID: 24999490 Free PMC article. - Visual pattern mining in histology image collections using bag of features.
Cruz-Roa A, Caicedo JC, González FA. Cruz-Roa A, et al. Artif Intell Med. 2011 Jun;52(2):91-106. doi: 10.1016/j.artmed.2011.04.010. Epub 2011 Jun 12. Artif Intell Med. 2011. PMID: 21664806 - Adapting content-based image retrieval techniques for the semantic annotation of medical images.
Kumar A, Dyer S, Kim J, Li C, Leong PH, Fulham M, Feng D. Kumar A, et al. Comput Med Imaging Graph. 2016 Apr;49:37-45. doi: 10.1016/j.compmedimag.2016.01.001. Epub 2016 Feb 4. Comput Med Imaging Graph. 2016. PMID: 26890880 - Semantic sparse recoding of visual content for image applications.
Lu Z, Han P, Wang L, Wen JR. Lu Z, et al. IEEE Trans Image Process. 2015 Jan;24(1):176-88. doi: 10.1109/TIP.2014.2375641. Epub 2014 Nov 26. IEEE Trans Image Process. 2015. PMID: 25438314 - Automatic medical image annotation and keyword-based image retrieval using relevance feedback.
Ko BC, Lee J, Nam JY. Ko BC, et al. J Digit Imaging. 2012 Aug;25(4):454-65. doi: 10.1007/s10278-011-9443-5. J Digit Imaging. 2012. PMID: 22193754 Free PMC article. Review.
Cited by
- High-throughput adaptive sampling for whole-slide histopathology image analysis (HASHI) via convolutional neural networks: Application to invasive breast cancer detection.
Cruz-Roa A, Gilmore H, Basavanhally A, Feldman M, Ganesan S, Shih N, Tomaszewski J, Madabhushi A, González F. Cruz-Roa A, et al. PLoS One. 2018 May 24;13(5):e0196828. doi: 10.1371/journal.pone.0196828. eCollection 2018. PLoS One. 2018. PMID: 29795581 Free PMC article. - Incorporating External Information in Tissue Subtyping: A Topic Modeling Approach.
Saeedi A, Yadollahpour P, Singla S, Pollack B, Wells W, Sciurba F, Batmanghelich K. Saeedi A, et al. Proc Mach Learn Res. 2021;149:478-505. Proc Mach Learn Res. 2021. PMID: 35098143 Free PMC article. - Latent representation learning in biology and translational medicine.
Kopf A, Claassen M. Kopf A, et al. Patterns (N Y). 2021 Mar 12;2(3):100198. doi: 10.1016/j.patter.2021.100198. eCollection 2021 Mar 12. Patterns (N Y). 2021. PMID: 33748792 Free PMC article. Review. - An alternative reference space for H&E color normalization.
Zarella MD, Yeoh C, Breen DE, Garcia FU. Zarella MD, et al. PLoS One. 2017 Mar 29;12(3):e0174489. doi: 10.1371/journal.pone.0174489. eCollection 2017. PLoS One. 2017. PMID: 28355298 Free PMC article.
References
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2006;445:168–76. - PubMed
- Kvilekval K, Fedorov D, Obara B, Singh A, Manjunath BS. Bisque: A platform for bioimage analysis and management. Bioinformatics. 2010;26:544–52. - PubMed