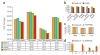Template-based protein structure modeling using the RaptorX web server - PubMed (original) (raw)
Template-based protein structure modeling using the RaptorX web server
Morten Källberg et al. Nat Protoc. 2012.
Abstract
A key challenge of modern biology is to uncover the functional role of the protein entities that compose cellular proteomes. To this end, the availability of reliable three-dimensional atomic models of proteins is often crucial. This protocol presents a community-wide web-based method using RaptorX (http://raptorx.uchicago.edu/) for protein secondary structure prediction, template-based tertiary structure modeling, alignment quality assessment and sophisticated probabilistic alignment sampling. RaptorX distinguishes itself from other servers by the quality of the alignment between a target sequence and one or multiple distantly related template proteins (especially those with sparse sequence profiles) and by a novel nonlinear scoring function and a probabilistic-consistency algorithm. Consequently, RaptorX delivers high-quality structural models for many targets with only remote templates. At present, it takes RaptorX ~35 min to finish processing a sequence of 200 amino acids. Since its official release in August 2011, RaptorX has processed ~6,000 sequences submitted by ~1,600 users from around the world.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Figures
Figure 1
Performance assessment of core prediction modules in the RaptorX server. (a) Comparison of structure prediction performance by global distance test (GDT) score for RaptorX and three other publicly available protocols on the CASP9 targets. Performance is compared in four categories: All CASP9 targets, template-based modeling (TBM) targets, hard TBM targets and multidomain targets. (b) Performance comparison for domain parsing between RaptorX and DoBo. Metrics are given for overall performance, and performance on single-domain and multidomain CASP9 target proteins. Specifically, accuracy is the overall proportion of both single-domain and multidomain proteins identified correctly; single (multi) recall is the fraction of single-domain (multidomain) proteins that are predicted; single (multi) precision is the fraction of correctly predicted single-domain (multidomain) proteins among all the predictions. A multidomain protein is correctly predicted only if its domain boundaries are correctly identified. (c) Performance comparison between RaptorX and PSIPRED for secondary structure prediction. The accuracies achieved for three-state prediction (helix, sheet and coil) are compared. (d) Performance comparison between RaptorX and SSPRO8 for eight-state secondary structure prediction. The accuracies achieved for eight-state prediction for the classes H, G, I, E, B, T, S, L (using SSPRO8 nomenclature) are compared.
Figure 2
Workflow used by the RaptorX server. Outline of the three modeling tasks users can accomplish using the RaptorX server, namely tertiary structure prediction, custom alignment and secondary structure prediction. For each stage, details of the computation and approximate completion time for a 250-residue target sequence are given (for threading; the indicated time is for a full template library scan). Blue boxes indicate mandatory stages, green boxes indicate optional stages and gray boxes indicate the resulting output. The blue, red and yellow directed paths indicate the flow for structure prediction, custom alignment jobs and secondary structure prediction, respectively. Dashed and solid paths indicate that the subsequent steps are optional and required, respectively. n/a, not applicable.
Figure 3
Job-listing interface. Selecting ‘My jobs’ displays this job overview for the user’s account, which gives the status of each prediction in the job along with overall information of the predictions being done for each sequence submitted.
Figure 4
Secondary structure result interface. The numbered labels indicate the location of the following screen features: (1) tabs for switching between the three-state and eight-state prediction; (2) hovering over a residue will give detailed statistics on the secondary-state distribution; (3) the status: a current running time of the job; (4) a download link for the prediction results; and (5) a color-code legend for secondary structure diagram.
Figure 5
Tertiary structure result interface. The numbered labels indicate the location of the following screen features: (1) the rank of currently selected model; (2) the quality score of the model; (3) the PDB IDs for the set templates used for modeling; (4) a drop-down menu for selecting alternative structure models; (5) tabs for switching between structure prediction, function annotation and BLAST output; (6) interactive viewer displaying the currently selected model structure; (7) menu for controlling the interactive viewer; (8) alignment used for structure modeling; (9) indication of the status: a current running time of the job; (10) download links for prediction results; and (11) a user guide for the interactive structure viewer.
Figure 6
Disorder prediction result display. Graphics comparable to those described for the secondary structure result interface (Fig. 4) are used to visualize the probability that a given residue is either in a disorder segment (marked in red) or nondisorder segment (marked in blue). Again, hovering over a residue will give detailed statistics on the disorder prediction, whereas the right-hand side shows the status of the job with a download link for the disorder prediction results and a color-code legend for the disorder prediction diagram.
Figure 7
Custom alignment result interface. The numbered labels indicate the location of the following screen features: (1) a drop-down menu for switching between alternative alignments; (2) the alignment between target sequence and template; (3) indication of the status: a current running time of the job; (4) a link for download of the prediction result; and (5) a legend indicating the alignment color coding.
Figure 8
Domain parsing result display. If multiple domains are found, the domain parsing results outline the span of each segment, the Pfam family it is predicted to belong to, a confidence measure (E value) for the domain assignment and a possible functional annotation of domain region.
Similar articles
- RaptorX server: a resource for template-based protein structure modeling.
Källberg M, Margaryan G, Wang S, Ma J, Xu J. Källberg M, et al. Methods Mol Biol. 2014;1137:17-27. doi: 10.1007/978-1-4939-0366-5_2. Methods Mol Biol. 2014. PMID: 24573471 - RaptorX: exploiting structure information for protein alignment by statistical inference.
Peng J, Xu J. Peng J, et al. Proteins. 2011;79 Suppl 10(Suppl 10):161-71. doi: 10.1002/prot.23175. Epub 2011 Oct 11. Proteins. 2011. PMID: 21987485 Free PMC article. - An Improved Integration of Template-Based and Template-Free Protein Structure Modeling Methods and its Assessment in CASP11.
Li J, Adhikari B, Cheng J. Li J, et al. Protein Pept Lett. 2015;22(7):586-93. doi: 10.2174/0929866522666150520145717. Protein Pept Lett. 2015. PMID: 25990081 Free PMC article. - Computational Methods for the Elucidation of Protein Structure and Interactions.
Edmunds NS, McGuffin LJ. Edmunds NS, et al. Methods Mol Biol. 2021;2305:23-52. doi: 10.1007/978-1-0716-1406-8_2. Methods Mol Biol. 2021. PMID: 33950383 Review. - Sequence comparison and protein structure prediction.
Dunbrack RL Jr. Dunbrack RL Jr. Curr Opin Struct Biol. 2006 Jun;16(3):374-84. doi: 10.1016/j.sbi.2006.05.006. Epub 2006 May 19. Curr Opin Struct Biol. 2006. PMID: 16713709 Review.
Cited by
- Predictive immunoinformatics reveal promising safety and anti-onchocerciasis protective immune response profiles to vaccine candidates (Ov-RAL-2 and Ov-103) in anticipation of phase I clinical trials.
Nebangwa DN, Shey RA, Shadrack DM, Shintouo CM, Yaah NE, Yengo BN, Efeti MT, Gwei KY, Fomekong DBA, Nchanji GT, Lemoge AA, Ntie-Kang F, Ghogomu SM. Nebangwa DN, et al. PLoS One. 2024 Oct 21;19(10):e0312315. doi: 10.1371/journal.pone.0312315. eCollection 2024. PLoS One. 2024. PMID: 39432476 Free PMC article. - Putative pseudolysogeny-dependent phage gene implicated in the superinfection resistance of Cutibacterium acnes.
Wottrich S, Mendonca S, Safarpour C, Nguyen C, Marinelli LJ, Hancock SP, Modlin RL, Parker JM. Wottrich S, et al. Microbiome Res Rep. 2024 Apr 18;3(3):27. doi: 10.20517/mrr.2023.42. eCollection 2024. Microbiome Res Rep. 2024. PMID: 39421248 Free PMC article. - Computational targeting of iron uptake proteins in Covid-19 induced mucormycosis to identify inhibitors via molecular dynamics, molecular mechanics and density function theory studies.
Sen M, Priyanka BM, Anusha D, Puneetha S, Setlur AS, Karunakaran C, Tandur A, Prashant CS, Niranjan V. Sen M, et al. In Silico Pharmacol. 2024 Sep 29;12(2):90. doi: 10.1007/s40203-024-00264-7. eCollection 2024. In Silico Pharmacol. 2024. PMID: 39355758 - Structure of the human TIP60-C histone exchange and acetyltransferase complex.
Li C, Smirnova E, Schnitzler C, Crucifix C, Concordet JP, Brion A, Poterszman A, Schultz P, Papai G, Ben-Shem A. Li C, et al. Nature. 2024 Sep 11. doi: 10.1038/s41586-024-08011-w. Online ahead of print. Nature. 2024. PMID: 39260417 - Web of venom: exploration of big data resources in animal toxin research.
Zancolli G, von Reumont BM, Anderluh G, Caliskan F, Chiusano ML, Fröhlich J, Hapeshi E, Hempel BF, Ikonomopoulou MP, Jungo F, Marchot P, de Farias TM, Modica MV, Moran Y, Nalbantsoy A, Procházka J, Tarallo A, Tonello F, Vitorino R, Zammit ML, Antunes A. Zancolli G, et al. Gigascience. 2024 Jan 2;13:giae054. doi: 10.1093/gigascience/giae054. Gigascience. 2024. PMID: 39250076 Free PMC article.
References
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources