Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma - PubMed (original) (raw)
. 2012 Aug 27;209(9):1553-65.
doi: 10.1084/jem.20120910. Epub 2012 Aug 13.
Thirunavukkarasu Velusamy, Bryan L Betz, Lili Zhao, Helmut G Weigelin, Mark Y Chiang, David R Huebner-Chan, Nathanael G Bailey, David T Yang, Govind Bhagat, Roberto N Miranda, David W Bahler, L Jeffrey Medeiros, Megan S Lim, Kojo S J Elenitoba-Johnson
Affiliations
- PMID: 22891276
- PMCID: PMC3428949
- DOI: 10.1084/jem.20120910
Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma
Mark J Kiel et al. J Exp Med. 2012.
Abstract
Splenic marginal zone lymphoma (SMZL), the most common primary lymphoma of spleen, is poorly understood at the genetic level. In this study, using whole-genome DNA sequencing (WGS) and confirmation by Sanger sequencing, we observed mutations identified in several genes not previously known to be recurrently altered in SMZL. In particular, we identified recurrent somatic gain-of-function mutations in NOTCH2, a gene encoding a protein required for marginal zone B cell development, in 25 of 99 (∼25%) cases of SMZL and in 1 of 19 (∼5%) cases of nonsplenic MZLs. These mutations clustered near the C-terminal proline/glutamate/serine/threonine (PEST)-rich domain, resulting in protein truncation or, rarely, were nonsynonymous substitutions affecting the extracellular heterodimerization domain (HD). NOTCH2 mutations were not present in other B cell lymphomas and leukemias, such as chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL; n = 15), mantle cell lymphoma (MCL; n = 15), low-grade follicular lymphoma (FL; n = 44), hairy cell leukemia (HCL; n = 15), and reactive lymphoid hyperplasia (n = 14). NOTCH2 mutations were associated with adverse clinical outcomes (relapse, histological transformation, and/or death) among SMZL patients (P = 0.002). These results suggest that NOTCH2 mutations play a role in the pathogenesis and progression of SMZL and are associated with a poor prognosis.
Figures
Figure 1.
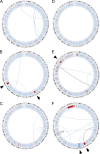
Structural alterations in index SMZL cases. Circos diagrams of genomic complexity identified in six index SMZL cases. Outer data track represents the relative sequencing coverage of chromosomal regions normalized to publicly available genome sequencing data of 49 healthy individuals corresponding to ploidy at these regions. Light blue indicates region of euploidy, dark blue indicates region of chromosomal gain and red indicates region of chromosomal loss. Arrows indicate deletion portions of the long arm of chromosome 7 (del7q) in two of the six index genomes. Arrowheads indicate other regions of chromosomal loss or gain. Inner data tracks represent large structural alterations between spatial distinct genomic regions affecting the coding regions of one or more genes. Light blue lines indicate a single gene involved in structural alteration. Black lines indicate two genes involved in structural alteration. The three index genomes with mutations in NOTCH2 are shown in A–C.
Figure 2.
WGS identifies NOTCH2 mutations in SMZL. (A and B) A representative case of SMZL with typical histopathological features of SMZL at low and high power (Bars: (A) 400 µm; (B) 50 µm) including expansion of pale staining marginal zones surrounding splenic follicles in a biphasic pattern. (C–E) Reverse complement sequence reads (Read Alignment) mapped to the reference genome (Reference Sequence) from three index samples with mutations in NOTCH2 (boxed) with deviations from reference genome highlighted in blue. Bottom panels show Sanger sequencing electropherograms confirming mutations in the index cases (SMZL) and the absence of the mutations in matched normal constitutional tissue (Germline). One frameshift p.I2304fsX9 mutation and two nonsense p.R2400X mutations were identified in three patients among the six index cases (arrows). The total number of reads containing the indicated mutation compared with the total number of reads mapping to this region is shown (C, 11/28; D, 16/44; E, 11/90). Genome sequencing was performed once for each index case. Sanger sequencing confirmation of somatic acquisition was performed in at least two independent replicates.
Figure 3.
Discovery, validation, and specificity assessment of NOTCH2 mutations in SMZL and other B cell lymphomas. A summary of the experimental design and results illustrates initial NOTCH2 mutation discovery in three of six index SMZL cases through WGS, all of which were confirmed as somatic mutations by traditional Sanger sequencing. Sanger sequencing of the C-terminal of NOTCH2 comprising exons 25–34 was performed in 93 additional SMZL cases comprising the validation cohort to establish the rate of recurrence of NOTCH2 mutations in SMZL. In total, 22 additional SMZL cases harbored mutations in the HD and PEST domains of NOTCH2. To assess the specificity of NOTCH2 mutations among other abnormal B cell proliferations, Sanger sequencing of NOTCH2 regions with recurrent mutations identified in discovery and validation samples of SMZL (exons 26, 27 and 34) was performed for 19 cases of nonsplenic marginal zone lymphoma (MALT-L), 14 cases of RLH, 15 chronic lymphocytic lymphoma (CLL/SLL), 44 low-grade follicular cell lymphoma (grade 1–2; FL), 15 HCL, and 15 MCL. To assess the consequences of NOTCH2 mutation on disease characteristics, clinical data were collected on a total of 46–53 SMZL cases including 11–12 cases with NOTCH2 mutation.
Figure 4.
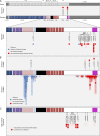
NOTCH2 mutations in SMZL. (A, top) The 34 exons of NOTCH2 are shown as gray boxes flanked by the 5′- and 3′-untranslated (UTR) regions of exons 1 and 34, respectively, above the protein domain structure of NOTCH2 including 36 epidermal growth factor-like repeats (EGFR, blue; mediates ligand binding), three LNR domains (purple; prevents ligand-independent activation), the HD (pink; prevents ligand-independent activation), a single-pass transmembrane region (TM, light blue), RBP-Jκ–associated module (RAM, black; required for NOTCH signaling), six ankyrin repeats (AR, red; bind the CSL transcription factor), the transactivation domain (TAD, white), and the proline-, glutamate-, serine- and threonine-rich domain (PEST, magenta). (A, Middle) Three mutations in the TAD and the PEST domain downstream of the AR region were identified in the SMZL discovery cohort. (A, Lower) Targeted Sanger sequencing of the SMZL validation cohort uncovered the same as well as additional missense (blue triangles), nonsense and frameshift (red circles) mutations in the HD, TAD and PEST domains. Constitutional tissue from a total of 19 patients confirmed somatic acquisition of these mutations in all samples (solid symbols). Sanger sequencing confirmation was performed in at least two independent replicates. (B) The locations of mutations in hematolymphoid malignancies in the NOTCH2 (7 total; top) and NOTCH1 (>800 total; bottom) genes in the COSMIC database are displayed adjacent to NOTCH2 and NOTCH1 amino acid sequence alignment. Mutations within both genes cluster in the HD and TAD/PEST domains. Specific amino acid residues and the predicted consequence of all NOTCH2 mutations in COSMIC are also displayed. (C) Mutations in HCS are confined to the C-terminal of the NOTCH2 protein and cluster within the TAD and PEST domains. The p.R2400X mutation seen in 9 cases of SMZL is also present in at least one case of HCS as an inherited mutation (open symbol). De novo mutations are displayed as solid symbols.
Figure 5.
Sanger sequencing identification of NOTCH2 mutations in SMZL validation cohort. Sanger sequencing results for each _NOTCH2_-mutated sample are shown. Arrows indicate site of nucleotide change. Amino acid change predicted for each mutation is indicated below sample label. Traces shown are representative of at least two independent amplification and sequencing reactions.
Figure 6.
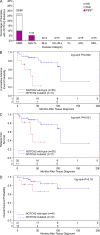
Impact of NOTCH2 mutations on clinical outcome in SMZL. (A) Frequency of NOTCH2 mutations in SMZL, MALT-L, and other B cell proliferative disorders divided among the different domains of the NOTCH2 protein (color corresponds to domain in which mutations were located). For each separate disease, the number of samples with NOTCH2 mutations compared with total number of samples analyzed is displayed above each bar. (B) Cumulative probability of relapse, transformation or death from time of tissue diagnosis for patients with _NOTCH2_-mutated (red lines) and _NOTCH2_-wild-type (blue lines) SMZL. (C) Relapse-free survival from tissue diagnosis. (D) Overall survival from tissue diagnosis. The total number of patients in each analysis is displayed along the x-axes of each graph.
Similar articles
- The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development.
Rossi D, Trifonov V, Fangazio M, Bruscaggin A, Rasi S, Spina V, Monti S, Vaisitti T, Arruga F, Famà R, Ciardullo C, Greco M, Cresta S, Piranda D, Holmes A, Fabbri G, Messina M, Rinaldi A, Wang J, Agostinelli C, Piccaluga PP, Lucioni M, Tabbò F, Serra R, Franceschetti S, Deambrogi C, Daniele G, Gattei V, Marasca R, Facchetti F, Arcaini L, Inghirami G, Bertoni F, Pileri SA, Deaglio S, Foà R, Dalla-Favera R, Pasqualucci L, Rabadan R, Gaidano G. Rossi D, et al. J Exp Med. 2012 Aug 27;209(9):1537-51. doi: 10.1084/jem.20120904. Epub 2012 Aug 13. J Exp Med. 2012. PMID: 22891273 Free PMC article. - KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype.
Clipson A, Wang M, de Leval L, Ashton-Key M, Wotherspoon A, Vassiliou G, Bolli N, Grove C, Moody S, Escudero-Ibarz L, Gundem G, Brugger K, Xue X, Mi E, Bench A, Scott M, Liu H, Follows G, Robles EF, Martinez-Climent JA, Oscier D, Watkins AJ, Du MQ. Clipson A, et al. Leukemia. 2015 May;29(5):1177-85. doi: 10.1038/leu.2014.330. Epub 2014 Nov 27. Leukemia. 2015. PMID: 25428260 - Whole exome sequencing of microdissected splenic marginal zone lymphoma: a study to discover novel tumor-specific mutations.
Peveling-Oberhag J, Wolters F, Döring C, Walter D, Sellmann L, Scholtysik R, Lucioni M, Schubach M, Paulli M, Biskup S, Zeuzem S, Küppers R, Hansmann ML. Peveling-Oberhag J, et al. BMC Cancer. 2015 Oct 24;15:773. doi: 10.1186/s12885-015-1766-z. BMC Cancer. 2015. PMID: 26498442 Free PMC article. - Splenic marginal zone lymphoma.
Piris MA, Onaindía A, Mollejo M. Piris MA, et al. Best Pract Res Clin Haematol. 2017 Mar-Jun;30(1-2):56-64. doi: 10.1016/j.beha.2016.09.005. Epub 2016 Nov 5. Best Pract Res Clin Haematol. 2017. PMID: 28288718 Review. - Genetic aberrations in small B-cell lymphomas and leukemias: molecular pathology, clinical relevance and therapeutic targets.
Bogusz AM, Bagg A. Bogusz AM, et al. Leuk Lymphoma. 2016 Sep;57(9):1991-2013. doi: 10.3109/10428194.2016.1173212. Epub 2016 Apr 27. Leuk Lymphoma. 2016. PMID: 27121112 Review.
Cited by
- First description of the t(3;17)(q27;q21)/IGF2BP2::LSM12 translocation in marginal zone lymphoma.
Diez-Feijóo R, Fernández-Rodríguez C, Lafuente M, García-Gisbert N, Ferrer A, Colomo L, Salido M, Salar A. Diez-Feijóo R, et al. Blood Adv. 2023 Jan 10;7(1):162-166. doi: 10.1182/bloodadvances.2022008393. Blood Adv. 2023. PMID: 36095303 Free PMC article. - Interferon regulatory factor 4 attenuates Notch signaling to suppress the development of chronic lymphocytic leukemia.
Shukla V, Shukla A, Joshi SS, Lu R. Shukla V, et al. Oncotarget. 2016 Jul 5;7(27):41081-41094. doi: 10.18632/oncotarget.9596. Oncotarget. 2016. PMID: 27232759 Free PMC article. - Whole-genome sequencing reveals oncogenic mutations in mycosis fungoides.
McGirt LY, Jia P, Baerenwald DA, Duszynski RJ, Dahlman KB, Zic JA, Zwerner JP, Hucks D, Dave U, Zhao Z, Eischen CM. McGirt LY, et al. Blood. 2015 Jul 23;126(4):508-19. doi: 10.1182/blood-2014-11-611194. Epub 2015 Jun 16. Blood. 2015. PMID: 26082451 Free PMC article. - Germline and somatic genetic variations of TNFAIP3 in lymphoma complicating primary Sjogren's syndrome.
Nocturne G, Boudaoud S, Miceli-Richard C, Viengchareun S, Lazure T, Nititham J, Taylor KE, Ma A, Busato F, Melki J, Lessard CJ, Sivils KL, Dubost JJ, Hachulla E, Gottenberg JE, Lombès M, Tost J, Criswell LA, Mariette X. Nocturne G, et al. Blood. 2013 Dec 12;122(25):4068-76. doi: 10.1182/blood-2013-05-503383. Epub 2013 Oct 24. Blood. 2013. PMID: 24159176 Free PMC article. Clinical Trial. - CBL-MZ is not a single biological entity: evidence from genomic analysis and prolonged clinical follow-up.
Parker H, McIver-Brown NR, Davis ZA, Parry M, Rose-Zerilli MJJ, Xochelli A, Gibson J, Walewska R, Strefford JC, Oscier DG. Parker H, et al. Blood Adv. 2018 May 22;2(10):1116-1119. doi: 10.1182/bloodadvances.2018019760. Blood Adv. 2018. PMID: 29773550 Free PMC article. No abstract available.
References
- Chacón J.I., Mollejo M., Muñoz E., Algara P., Mateo M., Lopez L., Andrade J., Carbonero I.G., Martínez B., Piris M.A., Cruz M.A. 2002. Splenic marginal zone lymphoma: clinical characteristics and prognostic factors in a series of 60 patients. Blood. 100:1648–1654 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous