The function of our microbiota: who is out there and what do they do? - PubMed (original) (raw)
Review
The function of our microbiota: who is out there and what do they do?
Noora Ottman et al. Front Cell Infect Microbiol. 2012.
Abstract
Current meta-omics developments provide a portal into the functional potential and activity of the intestinal microbiota. The comparative and functional meta-omics approaches have made it possible to get a molecular snap shot of microbial function at a certain time and place. To this end, metagenomics is a DNA-based approach, metatranscriptomics studies the total transcribed RNA, metaproteomics focuses on protein levels and metabolomics describes metabolic profiles. Notably, the metagenomic toolbox is rapidly expanding and has been instrumental in the generation of draft genome sequences of over 1000 human associated microorganisms as well as an astonishing 3.3 million unique microbial genes derived from the intestinal tract of over 100 European adults. Remarkably, it appeared that there are at least 3 clusters of co-occurring microbial species, termed enterotypes, that characterize the intestinal microbiota throughout various continents. The human intestinal microbial metagenome further revealed unique functions carried out in the intestinal environment and provided the basis for newly discovered mechanisms for signaling, vitamin production and glycan, amino-acid and xenobiotic metabolism. The activity and composition of the microbiota is affected by genetic background, age, diet, and health status of the host. In its turn the microbiota composition and activity influence host metabolism and disease development. Exemplified by the differences in microbiota composition and activity between breast- as compared to formula-fed babies, healthy and malnourished infants, elderly and centenarians as compared to youngsters, humans that are either lean or obese and healthy or suffering of inflammatory bowel diseases (IBD). In this review we will focus on our current understanding of the functionality of the human intestinal microbiota based on all available metagenome, metatranscriptome, and metaproteome results.
Keywords: functional metagenomics; human intestinal microbiota; metaproteomics; metatranscriptomics.
Figures
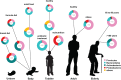
Figure 1
Human microbiota: onset and shaping through life stages and perturbations. The graph provides a global overview of the relative abundance of key phyla of the human microbiota composition in different stages of life. Measured by either 16S RNA or metagenomic approaches (DNA). Data arriving from: Babies breast- and formula-fed (Schwartz et al., 2012), baby solid food (Koenig et al., 2011), toddler antibiotic treatment (Koenig et al., 2011), toddler healthy or malnourished (Monira et al., 2011), adult, elderly, and centenarian healthy (Biagi et al., 2010), and adult obese (Zhang et al., 2009).
Similar articles
- Molecular ecological tools to decipher the role of our microbial mass in obesity.
Hermes GD, Zoetendal EG, Smidt H. Hermes GD, et al. Benef Microbes. 2015 Mar;6(1):61-81. doi: 10.3920/BM2014.0016. Benef Microbes. 2015. PMID: 24902956 Review. - Metagenomics and development of the gut microbiota in infants.
Vallès Y, Gosalbes MJ, de Vries LE, Abellán JJ, Francino MP. Vallès Y, et al. Clin Microbiol Infect. 2012 Jul;18 Suppl 4:21-6. doi: 10.1111/j.1469-0691.2012.03876.x. Clin Microbiol Infect. 2012. PMID: 22647043 - Application of metagenomics in the human gut microbiome.
Wang WL, Xu SY, Ren ZG, Tao L, Jiang JW, Zheng SS. Wang WL, et al. World J Gastroenterol. 2015 Jan 21;21(3):803-14. doi: 10.3748/wjg.v21.i3.803. World J Gastroenterol. 2015. PMID: 25624713 Free PMC article. Review. - Potential and active functions in the gut microbiota of a healthy human cohort.
Tanca A, Abbondio M, Palomba A, Fraumene C, Manghina V, Cucca F, Fiorillo E, Uzzau S. Tanca A, et al. Microbiome. 2017 Jul 14;5(1):79. doi: 10.1186/s40168-017-0293-3. Microbiome. 2017. PMID: 28709472 Free PMC article. - The human gut microbiome and its dysfunctions through the meta-omics prism.
Mondot S, Lepage P. Mondot S, et al. Ann N Y Acad Sci. 2016 May;1372(1):9-19. doi: 10.1111/nyas.13033. Epub 2016 Mar 4. Ann N Y Acad Sci. 2016. PMID: 26945826
Cited by
- Next generation probiotics for human health: An emerging perspective.
Jan T, Negi R, Sharma B, Kumar S, Singh S, Rai AK, Shreaz S, Rustagi S, Chaudhary N, Kaur T, Kour D, Sheikh MA, Kumar K, Yadav AN, Ahmed N. Jan T, et al. Heliyon. 2024 Aug 12;10(16):e35980. doi: 10.1016/j.heliyon.2024.e35980. eCollection 2024 Aug 30. Heliyon. 2024. PMID: 39229543 Free PMC article. Review. - The effects of stress on gut virome: Implications on infectious disease and systemic disorders.
Talarico F, Tilocca B, Spagnuolo R, Abenavoli L, Luzza F, Roncada P. Talarico F, et al. Microbiologyopen. 2024 Oct;13(5):e1434. doi: 10.1002/mbo3.1434. Microbiologyopen. 2024. PMID: 39311537 Free PMC article. Review. - Mango (Mangifera indica L.) Polyphenols: Anti-Inflammatory Intestinal Microbial Health Benefits, and Associated Mechanisms of Actions.
Kim H, Castellon-Chicas MJ, Arbizu S, Talcott ST, Drury NL, Smith S, Mertens-Talcott SU. Kim H, et al. Molecules. 2021 May 6;26(9):2732. doi: 10.3390/molecules26092732. Molecules. 2021. PMID: 34066494 Free PMC article. Review. - Intestinal microbiota and its role in irritable bowel syndrome (IBS).
Ohman L, Simrén M. Ohman L, et al. Curr Gastroenterol Rep. 2013 May;15(5):323. doi: 10.1007/s11894-013-0323-7. Curr Gastroenterol Rep. 2013. PMID: 23580243 Review. - Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study.
Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, Abe F, Osawa R. Odamaki T, et al. BMC Microbiol. 2016 May 25;16:90. doi: 10.1186/s12866-016-0708-5. BMC Microbiol. 2016. PMID: 27220822 Free PMC article.
References
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D. R., Fernandes G. R., Tap J., Bruls T., Batto J. M., Bertalan M., Borruel N., Casellas F., Fernandez L., Gautier L., Hansen T., Hattori M., Hayashi T., Kleerebezem M., Kurokawa K., Leclerc M., Levenez F., Manichanh C., Nielsen H. B., Nielsen T., Pons N., Poulain J., Qin J., Sicheritz-Ponten T., Tims S., Torrents D., Ugarte E., Zoetendal E. G., Wang J., Guarner F., Pedersen O., De Vos W. M., Brunak S., Dore J., Meta H. I. T. C., Antolin M., Artiguenave F., Blottiere H. M., Almeida M., Brechot C., Cara C., Chervaux C., Cultrone A., Delorme C., Denariaz G., Dervyn R., Foerstner K. U., Friss C., Van De Guchte M., Guedon E., Haimet F., Huber W., Van Hylckama-Vlieg J., Jamet A., Juste C., Kaci G., Knol J., Lakhdari O., Layec S., Le Roux K., Maguin E., Merieux A., Melo Minardi R., M'rini C., Muller J., Oozeer R., Parkhill J., Renault P., Rescigno M., Sanchez N., Sunagawa S., Torrejon A., Turner K., Vandemeulebrouck G., Varela E., Winogradsky Y., Zeller G., Weissenbach J., Ehrlich S. D., Bork P. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180 10.1038/nature09944 - DOI - PMC - PubMed
- Ben-Amor K., Heilig H., Smidt H., Vaughan E. E., Abee T., De Vos W. M. (2005). Genetic diversity of viable, injured, and dead fecal bacteria assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Appl. Environ. Microbiol. 71, 4679–4689 10.1128/AEM.71.8.4679-4689.2005 - DOI - PMC - PubMed
- Biagi E., Nylund L., Candela M., Ostan R., Bucci L., Pini E., Nikkila J., Monti D., Satokari R., Franceschi C., Brigidi P., De Vos W. (2010). Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE 5:e10667 10.1371/journal.pone.0010667 - DOI - PMC - PubMed
- Biasucci G., Benenati B., Morelli L., Bessi E., Boehm G. (2008). Cesarean delivery may affect the early biodiversity of intestinal bacteria. J. Nutr. 138, 1796S–1800S - PubMed
- Boesten R., Schuren F., Ben Amor K., Haarman M., Knol J., De Vos W. M. (2011). Bifidobacterium population analysis in the infant gut by direct mapping of genomic hybridization patterns: potential for monitoring temporal development and effects of dietary regimens. Microb. Biotechnol. 4, 417–427 10.1111/j.1751-7915.2010.00216.x - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources