Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus - PubMed (original) (raw)
Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus
Mang Shi et al. J Virol. 2012 Nov.
Abstract
PA-X is a fusion protein of influenza A virus encoded in part from a +1 frameshifted X open reading frame (X-ORF) in segment 3. We show that the X-ORFs of diverse influenza A viruses can be divided into two groups that differ in selection pressure and likely function, reflected in the presence of an internal stop codon and a change in synonymous diversity. Notably, truncated forms of PA-X evolved convergently in swine and dogs, suggesting a strong species-specific effect.
Figures
Fig 1
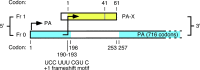
Influenza A virus segment 3 showing open reading frames for PA (in frame 0) and for the PA-X ORF (in frame 1), with the frameshift motif shown. Yellow shading indicates the region of the X-ORF that is translated; blue shading indicates structural domains of PA (1, 4, 10). The PA-X open reading frame encodes either 61 or 41 amino acids as indicated. Note that the X-ORF product lies largely within a linker region between the PA N- and C-terminal domains. (Reprinted from reference with permission of the publisher).
Fig 2
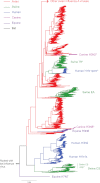
Phylogenetic tree of the full-length PA gene (frame 0) of influenza A virus showing the occurrence of the stop codon mutation at codon 42 in the X-ORF. Because of the very large size of the data set, this tree was inferred using a subsample of 3,281 PA sequences that differed by at least 0.005 nucleotide substitution per site. Clusters are colored according to host species, with those containing the stop codon mutation at codon 42 marked by asterisks. All horizontal branch lengths are drawn to a scale of nucleotide substitutions per site.
Similar articles
- An overlapping protein-coding region in influenza A virus segment 3 modulates the host response.
Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. Jagger BW, et al. Science. 2012 Jul 13;337(6091):199-204. doi: 10.1126/science.1222213. Epub 2012 Jun 28. Science. 2012. PMID: 22745253 Free PMC article. - Truncation of PA-X Contributes to Virulence and Transmission of H3N8 and H3N2 Canine Influenza Viruses in Dogs.
Liu L, Song S, Shen Y, Ma C, Wang T, Tong Q, Sun H, Pu J, Iqbal M, Liu J, Sun Y. Liu L, et al. J Virol. 2020 Jul 16;94(15):e00949-20. doi: 10.1128/JVI.00949-20. Print 2020 Jul 16. J Virol. 2020. PMID: 32461313 Free PMC article. - Virology. Frameshifting to PA-X influenza.
Yewdell JW, Ince WL. Yewdell JW, et al. Science. 2012 Jul 13;337(6091):164-5. doi: 10.1126/science.1225539. Science. 2012. PMID: 22798590 Free PMC article. - Translational termination-reinitiation in RNA viruses.
Powell ML. Powell ML. Biochem Soc Trans. 2010 Dec;38(6):1558-64. doi: 10.1042/BST0381558. Biochem Soc Trans. 2010. PMID: 21118126 Review. - [Classification and genome structure of influenza virus].
Maeda Y, Horimoto T, Kawaoka Y. Maeda Y, et al. Nihon Rinsho. 2003 Nov;61(11):1886-91. Nihon Rinsho. 2003. PMID: 14619426 Review. Japanese.
Cited by
- Influenza A Virus Protein PA-X Contributes to Viral Growth and Suppression of the Host Antiviral and Immune Responses.
Hayashi T, MacDonald LA, Takimoto T. Hayashi T, et al. J Virol. 2015 Jun;89(12):6442-52. doi: 10.1128/JVI.00319-15. Epub 2015 Apr 8. J Virol. 2015. PMID: 25855745 Free PMC article. - Bioinformatics studies of Influenza A hemagglutinin sequence data indicate recombination-like events leading to segment exchanges.
De A, Sarkar T, Nandy A. De A, et al. BMC Res Notes. 2016 Apr 15;9:222. doi: 10.1186/s13104-016-2017-3. BMC Res Notes. 2016. PMID: 27083561 Free PMC article. - PA-X: a key regulator of influenza A virus pathogenicity and host immune responses.
Hu J, Ma C, Liu X. Hu J, et al. Med Microbiol Immunol. 2018 Nov;207(5-6):255-269. doi: 10.1007/s00430-018-0548-z. Epub 2018 Jul 4. Med Microbiol Immunol. 2018. PMID: 29974232 Free PMC article. Review. - Pathogenic analysis of the pandemic 2009 H1N1 influenza A viruses in ferrets.
Tsuda Y, Weisend C, Martellaro C, Feldmann F, Haddock E. Tsuda Y, et al. J Vet Med Sci. 2017 Aug 18;79(8):1453-1460. doi: 10.1292/jvms.16-0619. Epub 2017 Jul 3. J Vet Med Sci. 2017. PMID: 28674309 Free PMC article. - An A14U Substitution in the 3' Noncoding Region of the M Segment of Viral RNA Supports Replication of Influenza Virus with an NS1 Deletion by Modulating Alternative Splicing of M Segment mRNAs.
Zheng M, Wang P, Song W, Lau SY, Liu S, Huang X, Mok BW, Liu YC, Chen Y, Yuen KY, Chen H. Zheng M, et al. J Virol. 2015 Oct;89(20):10273-85. doi: 10.1128/JVI.00919-15. Epub 2015 Jul 29. J Virol. 2015. PMID: 26223635 Free PMC article.
References
- Dias A, et al. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918 - PubMed
- He X, et al. 2008. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454:1123–1126 - PubMed
- Holmes EC, Lipman DJ, Zamarin D, Yewdell JW. 2006. Comment on “Large-scale sequence analysis of avian influenza isolates.” Science 313:1573 (Reply, 313:1573.) - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources