Loss of Lon1 in Arabidopsis changes the mitochondrial proteome leading to altered metabolite profiles and growth retardation without an accumulation of oxidative damage - PubMed (original) (raw)
Loss of Lon1 in Arabidopsis changes the mitochondrial proteome leading to altered metabolite profiles and growth retardation without an accumulation of oxidative damage
Cory Solheim et al. Plant Physiol. 2012 Nov.
Abstract
Lon1 is an ATP-dependent protease and chaperone located in the mitochondrial matrix in plants. Knockout in Arabidopsis (Arabidopsis thaliana) leads to a significant growth rate deficit in both roots and shoots and lowered activity of specific mitochondrial enzymes associated with respiratory metabolism. Analysis of the mitochondrial proteomes of two lon1 mutant alleles (lon1-1 and lon1-2) with different severities of phenotypes shows a common accumulation of several stress marker chaperones and lowered abundance of Complexes I, IV, and V of OXPHOS. Certain enzymes of the tricarboxylic acid (TCA) cycle are modified or accumulated, and TCA cycle bypasses were repressed rather than induced. While whole tissue respiratory rates were unaltered in roots and shoots, TCA cycle intermediate organic acids were depleted in leaf extracts in the day in lon1-1 and in both lon mutants at night. No significant evidence of broad steady-state oxidative damage to isolated mitochondrial samples could be found, but peptides from several specific proteins were more oxidized and selected functions were more debilitated in lon1-1. Collectively, the evidence suggests that loss of Lon1 significantly modifies respiratory function and plant performance by small but broad alterations in the mitochondrial proteome gained by subtly changing steady-state protein assembly, stability, and damage of a range of components that debilitate an anaplerotic role for mitochondria in cellular carbon metabolism.
Figures
Figure 1.
Growth habit, biomass, tissue respiratory capacity, and Lon1 abundance in lon1 mutants of Arabidopsis. A, Phenotype and dry weight of 3-week-old aerial tissues (n = 10; mean ±
se
). B, Root length of 15-d-old seedlings (n = 15; mean ±
se
). C, Shoot respiration rates (n = 13; mean ±
se
). D, Root respiration rates (n = 13; mean ±
se
). E, Average peptide abundance in MRM transitions (n = 6; mean ±
se
). Significant differences compared with the wild type are indicated: *P ≤ 0.05 and #P ≤ 0.01. FW, Fresh weight; WT, the wild type.
Figure 2.
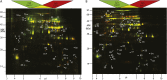
DIGE 2D IEF/SDS-PAGE of lon1 Arabidopsis mutants. Samples of mitochondria isolated from shoots of hydroponically grown lon1 mutants are compared with Col-0. A, lon1-1 (labeled with Cy3) compared with Col-0 (labeled with Cy5); yellow protein spots represent proteins of equal abundance between genotypes, while protein spots more abundant in lon1-1 mutant are green and protein spots more abundant in Col-0 are red. B, lon1-2 (labeled with Cy5) compared with Col-0 (labeled with Cy3); protein spots more abundant in lon1-2 mutant are red, and protein spots more abundant in Col-0 are green. Arrows indicate proteins unambiguously identified by MS; the numbers correlate with Table I. MW, Molecular weight.
Figure 3.
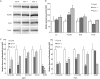
Mitochondrial respiratory complex activity, abundance, and subunit abundance of lon1 Arabidopsis mutants. A, Representative images of immunodetection of mitochondrial ETC subunits in SDS-PAGE separations of purified mitochondria. B, Relative protein amount of immunodetected mitochondrial ETC subunits (n = 3; mean ±
se
). C, Oxygen consumption of purified mitochondria (n ≥ 6; mean ±
se
). D, SDH activity of purified mitochondria (n ≥ 3; mean ±
se
). E, Relative abundance of mitochondrial ETC complexes determined from Coomassie-stained blue native-PAGE separations of isolated mitochondrial proteins (n = 3; mean ±
se
). Significant differences compared with the wild type (WT) indicated: *P ≤ 0.05 and #P ≤ 0.01.
Figure 4.
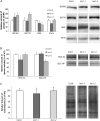
Immunodetection and global metabolite analysis of TCA cycle proteins and products in Arabidopsis lon1 mutants. A, Representative images of immunodetection of TCA cycle and related proteins in SDS-PAGE separations of purified mitochondria. B, Relative protein amount of immunodetected TCA cycle and related proteins (n = 3; mean ±
se
). C, Global levels of TCA cycle intermediates in light and dark isolated aerial tissues (n = 10; mean ±
se
). Significant differences compared with the wild type (WT) indicated: *P ≤ 0.05 and #P ≤ 0.01.
Figure 5.
Abundance of stress-related proteins and analysis of oxidation of Arabidopsis lon1 mutants. A, Relative protein amount of immunodetected stress-related proteins (n = 3; mean ±
se
) and representative images of immunodetection of stress-related proteins in SDS-PAGE separations of purified mitochondria. B, Relative amount of lipoate moieties in PDC-E2 and GDC-H protein of purified mitochondria (n = 3; mean ±
se
) and representative images of SDS-PAGE separated mitochondria immunodetected with antibodies to lipoic acid. C, Relative amount of carbonylation in purified mitochondria (n = 3; mean ±
se
) and representative lanes of SDS-PAGE separated mitochondria immunodetected with carbonyl antibodies. Significant differences compared with the wild type (WT) indicated: *P ≤ 0.05.
Figure 6.
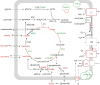
Diagram of the impact of Lon1 loss on mitochondrial metabolism. Red metabolites indicate organic acid pools decreasing in mutants (Fig. 4C). Red or green enzyme names indicate lower or higher protein abundance (Table I; Figs. 3–5). Red arrows indicate evidence of loss of enzyme activity (Figs. 2–4; Rigas et al., 2009a).
Similar articles
- Changes in specific protein degradation rates in Arabidopsis thaliana reveal multiple roles of Lon1 in mitochondrial protein homeostasis.
Li L, Nelson C, Fenske R, Trösch J, Pružinská A, Millar AH, Huang S. Li L, et al. Plant J. 2017 Feb;89(3):458-471. doi: 10.1111/tpj.13392. Epub 2017 Jan 7. Plant J. 2017. PMID: 27726214 - Role of Lon1 protease in post-germinative growth and maintenance of mitochondrial function in Arabidopsis thaliana.
Rigas S, Daras G, Laxa M, Marathias N, Fasseas C, Sweetlove LJ, Hatzopoulos P. Rigas S, et al. New Phytol. 2009;181(3):588-600. doi: 10.1111/j.1469-8137.2008.02701.x. Epub 2008 Dec 8. New Phytol. 2009. PMID: 19076295 - Lon1 Inactivation Downregulates Autophagic Flux and Brassinosteroid Biogenesis, Modulating Mitochondrial Proportion and Seed Development in Arabidopsis.
Song C, Hou Y, Li T, Liu Y, Wang XA, Qu W, Li L. Song C, et al. Int J Mol Sci. 2024 May 16;25(10):5425. doi: 10.3390/ijms25105425. Int J Mol Sci. 2024. PMID: 38791463 Free PMC article. - The multifaceted role of Lon proteolysis in seedling establishment and maintenance of plant organelle function: living from protein destruction.
Rigas S, Daras G, Tsitsekian D, Hatzopoulos P. Rigas S, et al. Physiol Plant. 2012 May;145(1):215-23. doi: 10.1111/j.1399-3054.2011.01537.x. Epub 2011 Dec 7. Physiol Plant. 2012. PMID: 22023720 Review. - The plant mitochondrial proteome.
Millar AH, Heazlewood JL, Kristensen BK, Braun HP, Møller IM. Millar AH, et al. Trends Plant Sci. 2005 Jan;10(1):36-43. doi: 10.1016/j.tplants.2004.12.002. Trends Plant Sci. 2005. PMID: 15642522 Review.
Cited by
- Lack of FTSH4 Protease Affects Protein Carbonylation, Mitochondrial Morphology, and Phospholipid Content in Mitochondria of Arabidopsis: New Insights into a Complex Interplay.
Smakowska E, Skibior-Blaszczyk R, Czarna M, Kolodziejczak M, Kwasniak-Owczarek M, Parys K, Funk C, Janska H. Smakowska E, et al. Plant Physiol. 2016 Aug;171(4):2516-35. doi: 10.1104/pp.16.00370. Epub 2016 Jun 13. Plant Physiol. 2016. PMID: 27297677 Free PMC article. - Mitochondrial CLPP2 Assists Coordination and Homeostasis of Respiratory Complexes.
Petereit J, Duncan O, Murcha MW, Fenske R, Cincu E, Cahn J, Pružinská A, Ivanova A, Kollipara L, Wortelkamp S, Sickmann A, Lee J, Lister R, Millar AH, Huang S. Petereit J, et al. Plant Physiol. 2020 Sep;184(1):148-164. doi: 10.1104/pp.20.00136. Epub 2020 Jun 22. Plant Physiol. 2020. PMID: 32571844 Free PMC article. - Glyoxalase I activity affects Arabidopsis sensitivity to ammonium nutrition.
Borysiuk K, Ostaszewska-Bugajska M, Kryzheuskaya K, Gardeström P, Szal B. Borysiuk K, et al. Plant Cell Rep. 2022 Dec;41(12):2393-2413. doi: 10.1007/s00299-022-02931-5. Epub 2022 Oct 15. Plant Cell Rep. 2022. PMID: 36242617 Free PMC article. - Is Autophagy Involved in Pepper Fruit Ripening?
López-Vidal O, Olmedilla A, Sandalio LM, Sevilla F, Jiménez A. López-Vidal O, et al. Cells. 2020 Jan 1;9(1):106. doi: 10.3390/cells9010106. Cells. 2020. PMID: 31906273 Free PMC article. - Evolution and significance of the Lon gene family in Arabidopsis organelle biogenesis and energy metabolism.
Rigas S, Daras G, Tsitsekian D, Alatzas A, Hatzopoulos P. Rigas S, et al. Front Plant Sci. 2014 Apr 11;5:145. doi: 10.3389/fpls.2014.00145. eCollection 2014. Front Plant Sci. 2014. PMID: 24782883 Free PMC article. Review.
References
- Abramoff MD, Magalhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophotonics International 11: 36–42
- Adam Z. (2000) Chloroplast proteases: possible regulators of gene expression? Biochimie 82: 647–654 - PubMed
- Barakat S, Pearce DA, Sherman F, Rapp WD. (1998) Maize contains a Lon protease gene that can partially complement a yeast pim1-deletion mutant. Plant Mol Biol 37: 141–154 - PubMed
- Bender T, Leidhold C, Ruppert T, Franken S, Voos W. (2010) The role of protein quality control in mitochondrial protein homeostasis under oxidative stress. Proteomics 10: 1426–1443 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases