Modulation of epigenetic targets for anticancer therapy: clinicopathological relevance, structural data and drug discovery perspectives - PubMed (original) (raw)
Review
Modulation of epigenetic targets for anticancer therapy: clinicopathological relevance, structural data and drug discovery perspectives
Federico Andreoli et al. Curr Pharm Des. 2013.
Free PMC article
Abstract
Research on cancer epigenetics has flourished in the last decade. Nevertheless growing evidence point on the importance to understand the mechanisms by which epigenetic changes regulate the genesis and progression of cancer growth. Several epigenetic targets have been discovered and are currently under validation for new anticancer therapies. Drug discovery approaches aiming to target these epigenetic enzymes with small-molecules inhibitors have produced the first pre-clinical and clinical outcomes and many other compounds are now entering the pipeline as new candidate epidrugs. The most studied targets can be ascribed to histone deacetylases and DNA methyltransferases, although several other classes of enzymes are able to operate post-translational modifications to histone tails are also likely to represent new frontiers for therapeutic interventions. By acknowledging that the field of cancer epigenetics is evolving with an impressive rate of new findings, with this review we aim to provide a current overview of pre-clinical applications of smallmolecules for cancer pathologies, combining them with the current knowledge of epigenetic targets in terms of available structural data and drug design perspectives.
Figures
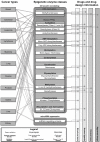
Fig. (1)
Connections between classes of epigenetic targets with cancer diseases and drug discovery information. Known ligands, clinical trials and approved drugs refer to cancer therapies with mechanisms of action directly related to epigenetic targets. Clinical trials and approved drugs have been recently reviewed in [46].
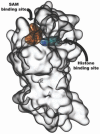
Fig. (2)
Depiction of SAM and histone binding sites of SETD8 methyltranferase. Assessing the mode of action implies a full understanding of the more appropriate mechanism of inhibition operated by small-molecules.
Similar articles
- Unravelling KDM4 histone demethylase inhibitors for cancer therapy.
Baby S, Gurukkala Valapil D, Shankaraiah N. Baby S, et al. Drug Discov Today. 2021 Aug;26(8):1841-1856. doi: 10.1016/j.drudis.2021.05.015. Epub 2021 May 26. Drug Discov Today. 2021. PMID: 34051367 Review. - Epigenetics and oncology.
Mummaneni P, Shord SS. Mummaneni P, et al. Pharmacotherapy. 2014 May;34(5):495-505. doi: 10.1002/phar.1408. Epub 2014 Mar 11. Pharmacotherapy. 2014. PMID: 24619798 Review. - Recent progress in the discovery of epigenetic inhibitors for the treatment of cancer.
Verma SK. Verma SK. Methods Mol Biol. 2015;1238:677-88. doi: 10.1007/978-1-4939-1804-1_35. Methods Mol Biol. 2015. PMID: 25421686 Review. - The Development of Epigenetics and Related Inhibitors for Targeted Drug Design in Cancer Therapy.
Liu N, Zhao R, Ma Y, Wang D, Yan C, Zhou D, Yin F, Li Z. Liu N, et al. Curr Top Med Chem. 2018;18(28):2380-2394. doi: 10.2174/1568026618666181115092623. Curr Top Med Chem. 2018. PMID: 30430946 Review. - DNA methyltransferases: emerging targets for the discovery of inhibitors as potent anticancer drugs.
Yu J, Xie T, Wang Z, Wang X, Zeng S, Kang Y, Hou T. Yu J, et al. Drug Discov Today. 2019 Dec;24(12):2323-2331. doi: 10.1016/j.drudis.2019.08.006. Epub 2019 Sep 5. Drug Discov Today. 2019. PMID: 31494187 Review.
Cited by
- Metabolic Response to XD14 Treatment in Human Breast Cancer Cell Line MCF-7.
Pan D, Kather M, Willmann L, Schlimpert M, Bauer C, Lagies S, Schmidtkunz K, Eisenhardt SU, Jung M, Günther S, Kammerer B. Pan D, et al. Int J Mol Sci. 2016 Oct 24;17(10):1772. doi: 10.3390/ijms17101772. Int J Mol Sci. 2016. PMID: 27783056 Free PMC article. - An RNA-seq-Based Expression Profiling of Radiation-Induced Esophageal Injury in a Rat Model.
Sun Z, Li J, Lin M, Zhang S, Luo J, Tang Y. Sun Z, et al. Dose Response. 2019 May 5;17(2):1559325819843373. doi: 10.1177/1559325819843373. eCollection 2019 Apr-Jun. Dose Response. 2019. PMID: 31105479 Free PMC article. - Transcriptome sequencing of microglial cells stimulated with TLR3 and TLR4 ligands.
Das A, Chai JC, Kim SH, Lee YS, Park KS, Jung KH, Chai YG. Das A, et al. BMC Genomics. 2015 Jul 10;16(1):517. doi: 10.1186/s12864-015-1728-5. BMC Genomics. 2015. PMID: 26159724 Free PMC article. - Targeting Histone Deacetylases in Diseases: Where Are We?
Benedetti R, Conte M, Altucci L. Benedetti R, et al. Antioxid Redox Signal. 2015 Jul 1;23(1):99-126. doi: 10.1089/ars.2013.5776. Epub 2014 Mar 6. Antioxid Redox Signal. 2015. PMID: 24382114 Free PMC article. Review. - MicroRNAs overexpressed in Crohn's disease and their interactions with mechanisms of epigenetic regulation explain novel aspects of Crohn's disease pathogenesis.
Fernández-Ponce C, Navarro Quiroz R, Díaz Perez A, Aroca Martinez G, Cadena Bonfanti A, Acosta Hoyos A, Gómez Escorcia L, Hernández Agudelo S, Orozco Sánchez C, Villarreal Camacho J, Atencio Ibarra L, Consuegra Machado J, Espinoza Garavito A, García-Cózar F, Navarro Quiroz E. Fernández-Ponce C, et al. Clin Epigenetics. 2021 Feb 18;13(1):39. doi: 10.1186/s13148-021-01022-8. Clin Epigenetics. 2021. PMID: 33602320 Free PMC article. Review.
References
- Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr opinion cell Biol. 2007;19(3 ):266–72. - PubMed
- Herceg Z, Ushijima T. Introduction: epigenetics and cancer. 1st ed. Elsevier Inc; 2010. - PubMed
- Baylin SB. Epigenetics and Cancer. The Molecular Basis of Cancer. 2008. pp. 57–65.
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4 ):693–705. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources