methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles - PubMed (original) (raw)
methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles
Altuna Akalin et al. Genome Biol. 2012.
Abstract
DNA methylation is a chemical modification of cytosine bases that is pivotal for gene regulation, cellular specification and cancer development. Here, we describe an R package, methylKit, that rapidly analyzes genome-wide cytosine epigenetic profiles from high-throughput methylation and hydroxymethylation sequencing experiments. methylKit includes functions for clustering, sample quality visualization, differential methylation analysis and annotation features, thus automating and simplifying many of the steps for discerning statistically significant bases or regions of DNA methylation. Finally, we demonstrate methylKit on breast cancer data, in which we find statistically significant regions of differential methylation and stratify tumor subtypes. methylKit is available at http://code.google.com/p/methylkit.
Figures
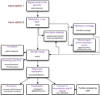
Figure 1
Flowchart of possible operations by methylKit. A summary of the most important_methylKit_ features is shown in a flow chart. It depicts the main features of _methylKit_and the sequential relationship between them. The functions that could be used for those features are also printed in the boxes.
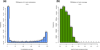
Figure 2
Descriptive statistics per sample. (a) Histogram of %methylation per cytosine for ER+ T47D sample. Most of the bases have either high or low methylation. (b) Histogram of read coverage per cytosine for ER+ T47D sample. ER+, estrogen receptor-alpha expressing.
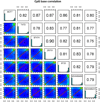
Figure 3
Scatter plots for sample pairs. Scatter plots of %methylation values for each pair in seven breast cancer cell lines. Numbers on upper right corner denote pair-wise Pearson's correlation scores. The histograms on the diagonal are %methylation histograms similar to Figure 2a for each sample.
Figure 4
Sample clustering. (a) Hierarchical clustering of seven breast cancer methylation profiles using 1-Pearson's correlation distance. (b) Principal Component Analysis (PCA) of seven breast cancer methylation profiles, plot shows principal component 1 and principal component 2 for each sample. Samples closer to each other in principal component space are similar in their methylation profiles.
Figure 5
Visualizing differential methylation events. (a) Horizontal bar plots show the number of hyper- and hypomethylation events per chromosome, as a percent of the sites with the minimum coverage and differential. By default this is a 25% change in methylation and all samples with 10X coverage. (b) Example of bedgraph file uploaded to UCSC browser. The bedraph file is for differentially methylated CpGs with at least a 25% difference and q-value <0.01. Hyper- and hypo-methylated bases are color coded. The bar heights correspond to % methylation difference between ER+ and ER- sets. ER+, estrogen receptor-alpha expressing; ER-, estrogen receptor-alpha non-expressing. UCSC, University of California Santa Cruz.
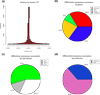
Figure 6
Annotation of differentially methylated CpGs. (a) Distance to TSS for differentially methylated CpGs are plotted from ER+ versus ER- analysis. (b) Pie chart showing percentages of differentially methylated CpGs on promoters, exons, introns and intergenic regions. (c) Pie chart showing percentages of differentially methylated CpGs on CpG islands, CpG island shores (defined as 2kb flanks of CpG islands) and other regions outside of shores and CpG islands. (d) Pie chart showing percentages of differentially methylated CpGs on enhancers and other regions. ER+, estrogen receptor-alpha expressing; ER-, estrogen receptor-alpha non-expressing, TSS, transcription start site.
Similar articles
- An optimized algorithm for detecting and annotating regional differential methylation.
Li S, Garrett-Bakelman FE, Akalin A, Zumbo P, Levine R, To BL, Lewis ID, Brown AL, D'Andrea RJ, Melnick A, Mason CE. Li S, et al. BMC Bioinformatics. 2013;14 Suppl 5(Suppl 5):S10. doi: 10.1186/1471-2105-14-S5-S10. Epub 2013 Apr 10. BMC Bioinformatics. 2013. PMID: 23735126 Free PMC article. - MethGo: a comprehensive tool for analyzing whole-genome bisulfite sequencing data.
Liao WW, Yen MR, Ju E, Hsu FM, Lam L, Chen PY. Liao WW, et al. BMC Genomics. 2015;16 Suppl 12(Suppl 12):S11. doi: 10.1186/1471-2164-16-S12-S11. Epub 2015 Dec 9. BMC Genomics. 2015. PMID: 26680022 Free PMC article. - Whole-Genome Bisulfite Sequencing for the Analysis of Genome-Wide DNA Methylation and Hydroxymethylation Patterns at Single-Nucleotide Resolution.
Kernaleguen M, Daviaud C, Shen Y, Bonnet E, Renault V, Deleuze JF, Mauger F, Tost J. Kernaleguen M, et al. Methods Mol Biol. 2018;1767:311-349. doi: 10.1007/978-1-4939-7774-1_18. Methods Mol Biol. 2018. PMID: 29524144 - Sequencing in High Definition Drives a Changing Worldview of the Epigenome.
Hodges E. Hodges E. Cold Spring Harb Perspect Med. 2019 Jun 3;9(6):a033076. doi: 10.1101/cshperspect.a033076. Cold Spring Harb Perspect Med. 2019. PMID: 30201789 Free PMC article. Review. - DNA methylation data by sequencing: experimental approaches and recommendations for tools and pipelines for data analysis.
Rauluseviciute I, Drabløs F, Rye MB. Rauluseviciute I, et al. Clin Epigenetics. 2019 Dec 12;11(1):193. doi: 10.1186/s13148-019-0795-x. Clin Epigenetics. 2019. PMID: 31831061 Free PMC article. Review.
Cited by
- DNA methylation correlates with transcriptional noise in response to elevated pCO2 in the eastern oyster (Crassostrea virginica).
Venkataraman YR, Huffmyer AS, White SJ, Downey-Wall A, Ashey J, Becker DM, Bengtsson Z, Putnam HM, Strand E, Rodríguez-Casariego JA, Wanamaker SA, Lotterhos KE, Roberts SB. Venkataraman YR, et al. Environ Epigenet. 2024 Sep 23;10(1):dvae018. doi: 10.1093/eep/dvae018. eCollection 2024. Environ Epigenet. 2024. PMID: 39534877 Free PMC article. - Nuclear localization of MTHFD2 is required for correct mitosis progression.
Pardo-Lorente N, Gkanogiannis A, Cozzuto L, Gañez Zapater A, Espinar L, Ghose R, Severino J, García-López L, Aydin RG, Martin L, Neguembor MV, Darai E, Cosma MP, Batlle-Morera L, Ponomarenko J, Sdelci S. Pardo-Lorente N, et al. Nat Commun. 2024 Nov 12;15(1):9529. doi: 10.1038/s41467-024-51847-z. Nat Commun. 2024. PMID: 39532843 Free PMC article. - LncRNA evolution and DNA methylation variation participate in photosynthesis pathways of distinct lineages of Populus.
Zhou J, Song F, He Y, Zhang W, Xiao L, Lu W, Li P, Quan M, Zhang D, Du Q. Zhou J, et al. For Res (Fayettev). 2023 Feb 6;3:3. doi: 10.48130/FR-2023-0003. eCollection 2023. For Res (Fayettev). 2023. PMID: 39526273 Free PMC article. - Changes in Gut Microbial Composition and DNA Methylation in Obese Patients with NAFLD After Bariatric Surgery.
Agodi A, Ojeda-Granados C, Maugeri A, Barchitta M, Coco O, Pezzino S, Magro G, Greca G, Latteri FS, Castorina S, Puleo S. Agodi A, et al. Int J Mol Sci. 2024 Oct 26;25(21):11510. doi: 10.3390/ijms252111510. Int J Mol Sci. 2024. PMID: 39519065 Free PMC article. - Gender-Specific DNA Methylation Profiles Associated with Adult Weight in Hezuo Pigs.
Jia R, Huang X, Yang J, Wang L, Li J, Li Y, Gun S, Yan Z, Wang P, Yang Q. Jia R, et al. Int J Mol Sci. 2024 Oct 25;25(21):11488. doi: 10.3390/ijms252111488. Int J Mol Sci. 2024. PMID: 39519040 Free PMC article.
References
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. - PubMed
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q-M, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical