Hypoxia-response element (HRE)-directed transcriptional regulation of the rat lysyl oxidase gene in response to cobalt and cadmium - PubMed (original) (raw)
Hypoxia-response element (HRE)-directed transcriptional regulation of the rat lysyl oxidase gene in response to cobalt and cadmium
Song Gao et al. Toxicol Sci. 2013 Apr.
Abstract
Lysyl oxidase (LO) catalyzes crosslink of collagen, elastin, and histone H1, stabilizing the extracellular matrix and cell nucleus. This enzyme displays dual functions for tumorigenesis, i.e., as a tumor suppressor inactivating the ras oncogene and as a tumor promoter enhancing malignant cell metastasis. To elucidate LO transcriptional regulation, we have cloned the 804 base pair region upstream of the translation start site (ATG) of the rat LO gene with the maximal promoter activity. Computer analysis indicated that at least four hypoxia-response element (HRE) consensuses (5'-ACGTG-3') exist in the cloned LO promoter. Treatment of rat lung fibroblasts (RFL6) with CoCl2 (Co, 10-100 μM), a chemical hypoxia reagent, enhanced LO mRNA expression and promoter activities. Overexpression of LO was associated with upregulation of hypoxia-inducible factor (HIF)-1α at mRNA levels in cobalt (Co)-treated cells. Thus, LO is a hypoxia-responsive gene. Dominant negative-HIF-1α inhibited LO promoter activities stimulated by Co. Electrophoretic mobility shift, oligonucleotide competition, and in vitro translated HIF-1α binding assays indicated that only one HRE mapped at -387/-383 relative to ATG was functionally active among four consensuses. Site-directed mutation of this HRE significantly diminished the Co-induced and LO promoter-directed expression of the reporter gene. Cadmium (Cd), an inducer of reactive oxygen species, inhibited HIF-1α mRNA expression and HIF-1α binding to the LO gene in Co-treated cells as revealed by RT-PCR and ChIP assays, respectively. Thus, modulation of the HRE activity by Co and Cd plays a critical role in LO gene transactivation.
Figures
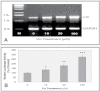
Fig. 1.
Co enhancement of LO mRNA levels (A) and promoter activities (B) in treated RFL6 cells. (A) Growth-arrested cells were incubated for 24h in 0.3% FBS/DMEM in the absence or presence of Co at indicated concentrations. Total RNA was extracted from cells using TRIzol reagent. LO mRNA levels in control and treated cells were determined by RT-PCR. GAPDH, an internal control; M, molecular ladder. Note: Densities of PCR-amplified gene fragments on the gel were measured with the 1D Scan software in this and below experiments. (B) Cells were cotransfected with the LO promoter-reporter construct (pLOProm 804) and phRL-TK vector, an internal control, then exposed to Co at indicated concentrations for 24h. Luciferase activity in each treatment was normalized to the internal control and expressed as % of the control without Co treatment. *p < 0.05, **p < 0.01, and ***p < 0.001 relative to control.
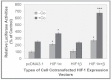
Fig. 2.
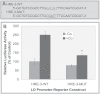
Co enhancement of the LO promoter activity stimulated by HIF-1 in treated RFL6 cells. Cells were cotransfected with the LO promoter-reporter construct (pLOProm 804) and different HIF-1 expression vectors as shown, as well as the internal control phRL-TK vector, then treated without or with 100µM Co for 24h. Luciferase activity in each treatment was normalized to the internal control and expressed as % of the control in which cells were cotransfected with the pcDNA3.1 basic vector without the HIF-1 cDNA insert. *p < 0.05 and ***p < 0.001 relative to control cells cotransfected with the pcDNA3.1 basic vector.
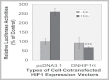
Fig. 3.
DN-HIF-1α inhibition of the LO promoter activity stimulated by Co in RFL6 cells. Cells were cotransfected with the LO promoter-reporter construct (pLOProm 804) and DN-HIF-1α expression vectors and the internal control phRL-TK vector, then treated without or with 100µM Co for 24h. Luciferase activity in each treatment was normalized to the internal control and expressed as % of the control in which cells were cotransfected with the pcDNA3.1 basic vector without the DN-HIF-1α cDNA insert. ***p < 0.001 relative to control cells cotransfected with the pcDNA3.1 basic vector without the DN-HIF-1α cDNA insert and treated with Co.
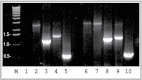
Fig. 4.
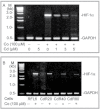
Association of upregulation of HIF-1α and LO at mRNA levels in RFL6 cells treated with Co. Growth-arrested cells were incubated for 24h in 0.3% FBS/DMEM in the absence (lanes 1–5) or presence (lanes 6–10) of 100µM Co. Total RNA was extracted from cells using TRIzol reagent. Transcript levels of HIF-1α (lanes 1 and 6), HIF-1β (lanes 2 and 7), DN-HIF-1α (lanes 3 and 8), LO (lanes 4 and 9), and GAPDH (lanes 5 and 10), an internal control, in control (lanes 1–5) and treated cells (lanes 6–10) were determined by RT-PCR. M, molecular ladder.
Fig. 5.
The schematic linear map of HRE consensuses in the cloned rat LO promoter. ATG, the translational start site; ACGTG or CACGT, the core HRE consensuses sequence; regions such as −387/−383 indicating the fragments between two nucleotide numbers using the first nucleotide preceding the ATG codon as −1. solid light blue line box, functionally active HRE consensuses; dash line boxes, nonfunction HRE consensuses; the green lines in the consensuses box, the HRE in the noncoding strand; and the light green lines in the coding strand; the red lines with orange boxes, coding regions.
Fig. 6.
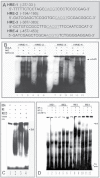
EMSA to determine functionally active LO HRE and nuclear protein binding. (A) Synthetic oligonucleotides containing HREs in the LO promoter region from −804 to −1 relative to ATG. (B) EMSA to assess nuclear protein binding. Synthetic oligonucleotides were end labeled with [γ-32P]ATP and incubated with nuclear protein isolated from control (NE), Co-treated cells (NECO), or bovine serum albumin (BSA), a negative control, as described in Materials and Methods section. After reaction, samples were subjected to native 6% polyacrylamide gel electrophoresis and visualized by exposure of the dried gel to Kodak film. (C) Competition EMSA. Cold synthetic human Epo HRE oligonucleotide with high affinity for the HRE (see Materials and Methods section) at 100-fold molecular excess was added 10min prior to addition of the radiolabeled probe. After reaction, gels were run as described above in B. (D) Supershift EMSA to assess DNA probe binding with in vitro transcription/translation HIF-1 proteins. Reaction mixtures contain 2 µg in vitro transcription/translation proteins, HIF-1α and HIF-1β each. Supershift reactions were run as described above in B with the exception that 2 µg of a specific antibody against HIF-1α was added 10min prior to addition of labeled probes.
Fig. 7.
Determination of the functional HRE by mutagenesis. (A) Mutation of the LO HRE −387/−383. The site-directed mutation of the HRE −387/−383 was performed with the QuikChange mutagenesis kit using pLOProm 804 as a basic. The core HRE 5′-CACGT-3′ was mutated to 5′-C_GAT_T-3′ labeled with gray color letters and underline. (B) Relative luciferase activities of LO promoter-reporter constructs. The wild or mutated LO promoter-reporter constructs each as shown above and the pRL-TK vector, an internal control, were transiently cotransfected into RFL6 cells. After transfection cells were treated with or without 100µM Co for 24h. Luciferase activity in each treatment was normalized to the internal control and expressed as % of the corresponding control. *p < 0.05 relative to the wild HRE vector–transfected control cells treated with Co.
Fig. 8.
HIF-1α binding to _cis_-element −387/−383 in RFL6 cells treated with Co, Cd, or both revealed by the ChIP assay. Growth-arrested RFL6 cells were treated without or with 100µM Co, 5µM Cd, or both for 24h (A and B). Control and CdR cells were treated without or with 100µM Co (C and D) for the same time. Cells at 2 × 106 for each group were processed for chromatin immunoprecipitation with an antibody against HIF-1α (A and C) or RNA-Poly II, a positive control (B and D). Using immunoprecipitated DNA as a template, the PCR with primer pairs, as shown in Materials and Methods section, amplified LO gene fragments containing the HRE −387/−383 with 150bp (A and C) and the GAPDH promoter fragment with 160bp (B and D). PCR products were analyzed on 2.2% agarose gels. Various treatments were indicated at the bottom on each gel. The left lane on each gel shows the DNA molecular ladder.
Fig. 9.
Cd inhibition of HIF-1α mRNA expression in RFL6 cells elicited by Co. Growth-arrested RFL6 cells were treated without or with 100µM Co in the presence of various concentration of Cd for 24h (A). Control and CdR cells were treated without or with 100µM Co for the same time (B). Total RNA was extracted from cells using TRIzol reagent. HIF-1α mRNA levels in control and treated cells were determined by RT-PCR. GAPDH, an internal control; M, molecular ladder.
Similar articles
- Up-Regulation of Neuronal Nitric Oxide Synthase Expression by Cobalt Chloride Through a HIF-1α Mechanism in Neuroblastoma Cells.
Li G, Zhao Y, Li Y, Lu J. Li G, et al. Neuromolecular Med. 2015 Dec;17(4):443-53. doi: 10.1007/s12017-015-8373-7. Epub 2015 Oct 12. Neuromolecular Med. 2015. PMID: 26458913 - Cloning and characterization of the rat lysyl oxidase gene promoter: identification of core promoter elements and functional nuclear factor I-binding sites.
Gao S, Zhao Y, Kong L, Toselli P, Chou IN, Stone P, Li W. Gao S, et al. J Biol Chem. 2007 Aug 31;282(35):25322-37. doi: 10.1074/jbc.M610108200. Epub 2007 Jun 27. J Biol Chem. 2007. PMID: 17597074 - Copper levels affect targeting of hypoxia-inducible factor 1α to the promoters of hypoxia-regulated genes.
Liu X, Zhang W, Wu Z, Yang Y, Kang YJ. Liu X, et al. J Biol Chem. 2018 Sep 21;293(38):14669-14677. doi: 10.1074/jbc.RA118.001764. Epub 2018 Aug 6. J Biol Chem. 2018. PMID: 30082314 Free PMC article. - Lysyl oxidase, a critical intra- and extra-cellular target in the lung for cigarette smoke pathogenesis.
Li W, Zhou J, Chen L, Luo Z, Zhao Y. Li W, et al. Int J Environ Res Public Health. 2011 Jan;8(1):161-84. doi: 10.3390/ijerph8010161. Epub 2011 Jan 19. Int J Environ Res Public Health. 2011. PMID: 21318022 Free PMC article. Review. - Hypoxia-inducible tumour-specific promoters as a dual-targeting transcriptional regulation system for cancer gene therapy.
Javan B, Shahbazi M. Javan B, et al. Ecancermedicalscience. 2017 Jul 6;11:751. doi: 10.3332/ecancer.2017.751. eCollection 2017. Ecancermedicalscience. 2017. PMID: 28798809 Free PMC article. Review.
Cited by
- Hypoxia-inducible factor 1α induces osteo/odontoblast differentiation of human dental pulp stem cells via Wnt/β-catenin transcriptional cofactor BCL9.
Orikasa S, Kawashima N, Tazawa K, Hashimoto K, Sunada-Nara K, Noda S, Fujii M, Akiyama T, Okiji T. Orikasa S, et al. Sci Rep. 2022 Jan 13;12(1):682. doi: 10.1038/s41598-021-04453-8. Sci Rep. 2022. PMID: 35027586 Free PMC article. - Conditioned CAR-T cells by hypoxia-inducible transcription amplification (HiTA) system significantly enhances systemic safety and retains antitumor efficacy.
He H, Liao Q, Zhao C, Zhu C, Feng M, Liu Z, Jiang L, Zhang L, Ding X, Yuan M, Zhang X, Xu J. He H, et al. J Immunother Cancer. 2021 Oct;9(10):e002755. doi: 10.1136/jitc-2021-002755. J Immunother Cancer. 2021. PMID: 34615704 Free PMC article. - Lysyl Oxidase Gene G473A Polymorphism and Cigarette Smoking in Association with a High Risk of Lung and Colorectal Cancers in a North Chinese Population.
Wang G, Shen Y, Cheng G, Bo H, Lin J, Zheng M, Li J, Zhao Y, Li W. Wang G, et al. Int J Environ Res Public Health. 2016 Jun 28;13(7):635. doi: 10.3390/ijerph13070635. Int J Environ Res Public Health. 2016. PMID: 27367711 Free PMC article. - Intrauterine Exposure to Cadmium Reduces HIF-1 DNA-Binding Ability in Rat Fetal Kidneys.
Jacobo-Estrada T, Cardenas-Gonzalez M, Santoyo-Sánchez MP, Thevenod F, Barbier O. Jacobo-Estrada T, et al. Toxics. 2018 Sep 3;6(3):53. doi: 10.3390/toxics6030053. Toxics. 2018. PMID: 30177602 Free PMC article. - Regulators of collagen crosslinking in developing and adult tendons.
Ellingson AJ, Pancheri NM, Schiele NR. Ellingson AJ, et al. Eur Cell Mater. 2022 Apr 5;43:130-152. doi: 10.22203/eCM.v043a11. Eur Cell Mater. 2022. PMID: 35380167 Free PMC article. Review.
References
- Ardyanto T. D., Osaki M., Tokuyasu N., Nagahama Y., Ito H. (2006). CoCl2-induced HIF-1alpha expression correlates with proliferation and apoptosis in MKN-1 cells: A possible role for the PI3K/Akt pathway. Int. J. Oncol. 29, 549–555 - PubMed
- Bruick R. K., McKnight S. L. (2001). A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294, 1337–1340 - PubMed
- Chen L. J., Zhao Y., Gao S., Chou I. N., Toselli P., Stone P., Li W. (2005). Downregulation of lysyl oxidase and upregulation of cellular thiols in rat fetal lung fibroblasts treated with cigarette smoke condensate. Toxicol. Sci. 83, 372–379 - PubMed
- Chun Y. S., Choi E., Kim G. T., Choi H., Kim C. H., Lee M. J., Kim M. S., Park J. W. (2000). Cadmium blocks hypoxia-inducible factor (HIF)-1-mediated response to hypoxia by stimulating the proteasome-dependent degradation of HIF-1alpha. Eur. J. Biochem. 267, 4198–4204 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources