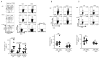B lymphocyte-induced maturation protein-1 contributes to intestinal mucosa homeostasis by limiting the number of IL-17-producing CD4+ T cells - PubMed (original) (raw)
B lymphocyte-induced maturation protein-1 contributes to intestinal mucosa homeostasis by limiting the number of IL-17-producing CD4+ T cells
Soofia Salehi et al. J Immunol. 2012.
Abstract
The transcription factor B lymphocyte-induced maturation protein-1 (Blimp-1) plays important roles in embryonic development and immunity. Blimp-1 is required for the differentiation of plasma cells, and mice with T cell-specific deletion of Blimp-1 (Blimp-1CKO mice) develop a fatal inflammatory response in the colon. Previous work demonstrated that lack of Blimp-1 in CD4(+) and CD8(+) T cells leads to intrinsic functional defects, but little is known about the functional role of Blimp-1 in regulating differentiation of Th cells in vivo and their contribution to the chronic intestinal inflammation observed in the Blimp1CKO mice. In this study, we show that Blimp-1 is required to restrain the production of the inflammatory cytokine IL-17 by Th cells in vivo. Blimp-1CKO mice have greater numbers of IL-17-producing TCRβ(+)CD4(+)cells in lymphoid organs and in the intestinal mucosa. The increase in IL-17-producing cells was not restored to normal levels in wild-type and Blimp-1CKO-mixed bone marrow chimeric mice, suggesting an intrinsic role for Blimp-1 in constraining the production of IL-17 in vivo. The observation that Blimp-1-deficient CD4(+) T cells are more prone to differentiate into IL-17(+)/IFN-γ(+) cells and cause severe colitis when transferred to Rag1-deficient mice provides further evidence that Blimp-1 represses IL-17 production. Analysis of Blimp-1 expression at the single cell level during Th differentiation reveals that Blimp-1 expression is induced in Th1 and Th2 but repressed by TGF-β in Th17 cells. Collectively, the results described here establish a new role for Blimp-1 in regulating IL-17 production in vivo.
Conflict of interest statement
The authors have no conflicting financial interests.
Figures
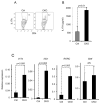
Figure 1. Accumulation of CD4+ TCRβ+ cells in the colon and increased amounts of IFNγ and IL17-producing cells in Blimp-1 CKO mice
A) Total numbers of mononuclear cells in the large intestines (LI: cecum, colon and rectum) lamina propria (LP) in 8–16 week-old Prdm1+/+CD4-CRE+/−(Ctrl) and _Prdm1_F/F CD4-CRE+/− (CKO) mice. Each symbol represents one animal (circles, Ctrl; triangles, CKO) and bars represent average for each group. Results shown are representative of 5 independent experiments. B) Percentages of TCRβ+CD4+ T cells (FACS plots) and total numbers of TCRβ+CD4+ T cells (chart) in LI-LP from Ctrl and Blimp-1CKO mice. C) Production of and IL17A (left) and IFNγ (right) measured by ELISA on supernatants of LI-LP cells stimulated with plate-bound anti-CD3 plus anti-CD28 for 24 hours. D) Intracellular staining (ICC) for lL17A and IFNγ (D, left plots) or IL17A and IL17F (D, right plots) and FACS analysis of CD4+TCRβ+ cells in mesenteric lymph nodes (mLN, top row) and spleen (SP, bottom row) from Ctrl and CKO mice. SP and mLN total cell suspensions were stimulated as in C) before analysis (BFA was added in the last 6 hours). Data shown are from gated TCRβ+CD4+ cells. E–F) combined results of the percentage (E) or absolute numbers (F) of IL17A or IFNγ+ CD4+ T cells in SP or mLN from 5–6 independent experiments for a total of 6–8 mice per group. G) Production of IL17A (ELISA) by cells from SP or mLN after 18 hours stimulation w/plate-bound anti-CD3 plus anti-CD28. Data show is the average and SEM from three different experiments with three to seven mice per group.
Figure 2. Increased expression of IL17 and Th17-signature genes in Blimp-1-deficient effector/memory cells
CD4+ antigen-experienced (CD44High) T cells were sorted from pooled SP and mLN from Ctrl or Blimp-1 CKO mice and stimulated in vitro with plate-bound anti-CD3 plus anti-CD28 and IL17 production was measured by ICC staining and FACS analysis at 6 hours (A) or in the supernatants by ELISA at 48 hours (B). Il17a, Il23r, RORC, and BATF steady state mRNA levels (C) were measured by qRT PCR in Ctrl and Blimp-1 CKO stimulated as in (A). Results shown in A-C are representative of two to three independent experiments with three to five mice/group. Bars show average and error bars represent SEM. Results in C are presented relative to 18S expression.
Figure 3. Increased production of IL17 by Blimp-1-deficient CD4+ T cells in mixed-bone marrow chimeric mice
Wild type CD45.1-irradiated mice received a total of ~4 × 106 HSC-enriched mixed BM cells from Ctrl (CD451.1) and Blimp-1CKO (CD451.2) mice. Eleven weeks after bone marrow cells transfer, mLN, SP and LI-LP cells were isolated from chimeric mice, stimulated as described in Fig. 1D and IL17 (A) production was analyzed in TCRβ+CD4+ cells in the CD45.2− (Ctrl) or CD45.2+ (Blimp1CKO) gates. B, Frequency of IFNγ-producing TCRβ+CD4+ cells from the MLN and SP from chimeric mice (stimulation and analysis done as in A). C) Frequency of Foxp3-expressing cells in the Ctrl and Blimp-1CKO compartments in the SP and mLN from chimeric mice (gated as shown in A). Plots (A–C) show compiled results from four to nine different chimeric mice.
Figure 4. Blimp-1 CKO CD4+ T cells differentiate into IL17 and IFNγ-double producer cells in vivo and cause severe colitis in Rag1−/− mice
Rag1−/− mice were injected i.p. with 4x105 naïve (CD44lowCD25−) CD4+ T cells sorted from Ctrl or Blimp-1 CKO mice and monitored for symptoms of colitis development and wasting disease, including body weight loss daily (A). Six to seven weeks after T cell transfer, recipient mice were euthanized and colons were collected and processed for H&E staining (B, 20 X magnifications) and histological analysis. Slides were analyzed blindly. Colitis score (C) was graded semi-quantitatively from 0–4, as previously described (35) each symbol represent one animal (the circles and triangles represent Ctrl and CKO, respectively). D) ICC staining and FACS analysis of cells from mLN from recipient mice 7 weeks after adoptive T cell transfer of Ctrl (circles) or Blimp1CKO (triangles) cells. mLN cells were stimulated in vitro for 24 hours with plate-bound αCD3 plus αCD28 before staining. FACS plots show cells in the TCRβ+CD4+ gate. E) Combined results (each symbol represent one animal, and bars represent average of five to six different experiments) of the percentage of IL17A+, IFNγ+, or IL17A+/IFNγ+ CD4+ T cells in the mLN (left panel), or LI-LP (right panel) for a total of six to seven mice per group. F) Absolute numbers of IL17A+, IFNγ+, or IL17A+/IFNγ+ CD4+ T cells in the mLN based on the percentages shown in E. G) Percentages of TCRβ+ CD4+Foxp3+ cells in the mLN (left) or LI-LP (right) of Rag−/− mice injected with Ctrl or Blimp-1 CKO 4x105 naïve [CD44lowCD25−GFP− (Foxp3−)] CD4+ T cells and analyzed six to seven weeks later. Data are representative of five (A–C) three (E–F) different experiments, with a total of four to seven mice per group.
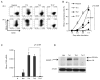
Figure 5. Blimp-1 is expressed in Th1 and Th2 but not Th17 cells
A) Blimp-1 expression as reported by YFP in Naïve CD4+ T cells sorted from YFP transgenic Blimp-1 reporter mice and stimulated under neutral (with IL2), Th1,Th2 or Th17 conditions for 3.5 (top row) or 7.5 (bottom row) days. FACS Plots shown are from the gated live, CD4+ cells in the well. B) Average percentages (and standard deviation) of YFP positive cells shown in A for five independent experiments, for a total of seven different mice. C) Steady state Blimp-1 mRNA at d7.5 in cells from experiment shown in A). D) Immunoblotting of total cell lysates obtained from cells stimulated as in A) for 5.5 days (Ntrl=Neutral), using antibodies to Blimp-1 or β-actin.
Figure 6. Blimp-1 expression is induced by Th1 and Th2-inducing cytokines but repressed by TGFβ in Th17-developing cells
A) Naïve CD4+ T cells from Blimp-1 reporter mice were stimulated with plate-bound anti-CD3 plus anti-CD28 alone or in the presence of the indicted cytokines and YFP (Blimp-1) expression was analyzed by FACS at d5. B) qRT-PCR analysis of Blimp-1 mRNA expression in the same cells from experiment shown in (A). C) Dose-dependent inhibition of Blimp-1 expression by TGFβ in Blimp-1 reporter naïve CD4+ T cells stimulated as indicated in the presence of different amounts of TGFβ for 5 days and then analyzed by FACS. D) qRT-PCR analysis of Blimp-1 mRNA expression in the same cells from experiment shown in (C). Data shown are from one experiment with pooled cells from three mice; similar results were obtained in 2 independent experiments.
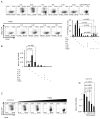
Figure 7. Blimp-1 binds to the murine Il17a gene in Th2 cells but it is not sufficient to repress IL17 production in Th17 cells
A) Chromatin Immunoprecipitation (ChIP) analysis of wild-type Th2 polarized cells re-stimulated with PMA and ionomycin, followed by immunoprecipitation of chromatin with anti-Blimp-1 or control antibody and quantitative PCR analysis of binding of Blimp-1 at different regions upstream (−3.3, −7.2 Kb ) or downstream (+44.9Kb) of the transcription start site of the murine Il17a gene or a negative control region in the SNAIL3 gene; results were normalized to those of the input chromatin, followed by the ratio of results obtained with anti-Blimp-1 antibody and control antibody (nonspecific background). Data are representative of three to four independent experiments (mean and SEM). B–C) Production of IL17 upon enforced expression of Blimp-1 in Th17 cells. Naïve CD4+ T cells were stimulated under Th17 conditions and transduced with retrovirus expressing GFP only (Ctrl RV) or GFP and Blimp-1 as a bi-cistronic message (Blimp-1 RV); cells were re-stimulated 3 (B) or 4 (C) days after transduction and analyzed for IL17A (top plots) or IL17F (bottom plots) production. Plots shown are from CD4+ (B) or CD4+GFP+(C) cells. Chart in C shows the average and SEM of three independent experiments.
Similar articles
- Egr-2 transcription factor is required for Blimp-1-mediated IL-10 production in IL-27-stimulated CD4+ T cells.
Iwasaki Y, Fujio K, Okamura T, Yanai A, Sumitomo S, Shoda H, Tamura T, Yoshida H, Charnay P, Yamamoto K. Iwasaki Y, et al. Eur J Immunol. 2013 Apr;43(4):1063-73. doi: 10.1002/eji.201242942. Epub 2013 Feb 26. Eur J Immunol. 2013. PMID: 23349024 - Blimp-1 attenuates Th1 differentiation by repression of ifng, tbx21, and bcl6 gene expression.
Cimmino L, Martins GA, Liao J, Magnusdottir E, Grunig G, Perez RK, Calame KL. Cimmino L, et al. J Immunol. 2008 Aug 15;181(4):2338-47. doi: 10.4049/jimmunol.181.4.2338. J Immunol. 2008. PMID: 18684923 - T-helper 17 and interleukin-17-producing lymphoid tissue inducer-like cells make different contributions to colitis in mice.
Ono Y, Kanai T, Sujino T, Nemoto Y, Kanai Y, Mikami Y, Hayashi A, Matsumoto A, Takaishi H, Ogata H, Matsuoka K, Hisamatsu T, Watanabe M, Hibi T. Ono Y, et al. Gastroenterology. 2012 Nov;143(5):1288-1297. doi: 10.1053/j.gastro.2012.07.108. Epub 2012 Jul 28. Gastroenterology. 2012. PMID: 22850180 - New insights into Blimp-1 in T lymphocytes: a divergent regulator of cell destiny and effector function.
Fu SH, Yeh LT, Chu CC, Yen BL, Sytwu HK. Fu SH, et al. J Biomed Sci. 2017 Jul 21;24(1):49. doi: 10.1186/s12929-017-0354-8. J Biomed Sci. 2017. PMID: 28732506 Free PMC article. Review. - Reciprocal IFN-gamma and TGF-beta responses regulate the occurrence of mucosal inflammation.
Strober W, Kelsall B, Fuss I, Marth T, Ludviksson B, Ehrhardt R, Neurath M. Strober W, et al. Immunol Today. 1997 Feb;18(2):61-4. doi: 10.1016/s0167-5699(97)01000-1. Immunol Today. 1997. PMID: 9057354 Review.
Cited by
- Blimp-1-Dependent IL-10 Production by Tr1 Cells Regulates TNF-Mediated Tissue Pathology.
Montes de Oca M, Kumar R, de Labastida Rivera F, Amante FH, Sheel M, Faleiro RJ, Bunn PT, Best SE, Beattie L, Ng SS, Edwards CL, Muller W, Cretney E, Nutt SL, Smyth MJ, Haque A, Hill GR, Sundar S, Kallies A, Engwerda CR. Montes de Oca M, et al. PLoS Pathog. 2016 Jan 14;12(1):e1005398. doi: 10.1371/journal.ppat.1005398. eCollection 2016 Jan. PLoS Pathog. 2016. PMID: 26765224 Free PMC article. - Genetic interactions between the DBL-1/BMP-like pathway and dpy body size-associated genes in Caenorhabditis elegans.
Lakdawala MF, Madhu B, Faure L, Vora M, Padgett RW, Gumienny TL. Lakdawala MF, et al. Mol Biol Cell. 2019 Dec 15;30(26):3151-3160. doi: 10.1091/mbc.E19-09-0500. Epub 2019 Nov 6. Mol Biol Cell. 2019. PMID: 31693440 Free PMC article. - The Lysine Methyltransferase G9a in Immune Cell Differentiation and Function.
Scheer S, Zaph C. Scheer S, et al. Front Immunol. 2017 Apr 11;8:429. doi: 10.3389/fimmu.2017.00429. eCollection 2017. Front Immunol. 2017. PMID: 28443098 Free PMC article. Review. - Regulation of CD4 T-cell differentiation and inflammation by repressive histone methylation.
Antignano F, Zaph C. Antignano F, et al. Immunol Cell Biol. 2015 Mar;93(3):245-52. doi: 10.1038/icb.2014.115. Epub 2015 Jan 13. Immunol Cell Biol. 2015. PMID: 25582341 Review. - Distinct roles for Blimp-1 in autoreactive CD4 T cells during priming and effector phase of autoimmune encephalomyelitis.
Aqel SI, Granitto MC, Nuro-Gyina PK, Pei W, Liu Y, Lovett-Racke AE, Racke MK, Yang Y. Aqel SI, et al. J Neuroimmunol. 2018 Dec 15;325:20-28. doi: 10.1016/j.jneuroim.2018.10.007. Epub 2018 Oct 15. J Neuroimmunol. 2018. PMID: 30366205 Free PMC article.
References
- Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. - PubMed
- Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI071087/AI/NIAID NIH HHS/United States
- R21 AI083948/AI/NIAID NIH HHS/United States
- P01 AI061093/AI/NIAID NIH HHS/United States
- AI083948-01/AI/NIAID NIH HHS/United States
- P01 DK046763/DK/NIDDK NIH HHS/United States
- R01 AI103542/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials