IGF-I 3' untranslated region: strain-specific polymorphisms and motifs regulating IGF-I in osteoblasts - PubMed (original) (raw)
IGF-I 3' untranslated region: strain-specific polymorphisms and motifs regulating IGF-I in osteoblasts
Spenser S Smith et al. Endocrinology. 2013 Jan.
Abstract
Reduced IGF-I is associated with low bone mass in humans and mice. C3H/He/J (C3H) mice have higher skeletal IGF-I and greater bone mass than C57BL/6J (B6). We hypothesized that strain-related genotypic differences in Igf1 affected skeletal function. The Igf1 coding region is nonpolymorphic, but its 3' untranslated region (UTR) is polymorphic between C3H and B6. Luciferase-Igf1 3' UTR reporter constructs showed that these polymorphic regions did not affect UTR function. IGF-I splice variants give rise to a common mature IGF-I peptide, but different E peptides. We identified two splice products, exon 4+6 (Ea) and exon 4+5+6 (Eb, mechano-growth factor) and found that their abundance was unchanged during osteoblastic differentiation. The Igf1 3' UTR encoded by exon 6 contains alternative polyadenylation sites. Proximal site use produces a short 3' UTR of approximately 195 bases, whereas distal site usage results in an approximately 6300-base UTR. Although Igf1 mRNA levels did not change during osteoblastic differentiation, distal polyadenylation site usage was increased in B6 cells but not in C3H. The resulting long Igf1 RNA isoform is less stable and has decreased translation efficiency, which may be one mechanism contributing to decreased IGF-I in B6 vs. C3H mice. Although the long UTR contains a conserved [GU](18) repeat, which is a positive regulator of UTR activity, it is also targeted by negative regulators, miR-29 and miR-365. These microRNAs are increased in B6 and C3H cells during osteoblastic differentiation. Differential expression of the long Igf1 3' UTR isoform may be a possible mechanism for enhanced IGF-I regulation in B6 vs. C3H mice.
Figures
Fig. 1.
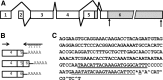
Polymorphisms in the Igf1 3′ UTR from B6 and C3H mice do not affect UTR function. A, Schematic representation of the Igf1 3′ UTR from exon 6 and the 3′ UTR fragments cloned into pMIR-REPORT Luciferase for functional analysis. Fragment 1 is nonpolymorphic. Fragments 2–4 are polymorphic, and strain-specific constructs were created. The proximal (open triangle) and distal (filled triangle) polyadenylation sites are indicated. The three polymorphic regions are denoted by arrows. B, Activity of luciferase-3′ UTR constructs in transiently transfected MC3T3 cells. All Igf1 3′ UTR constructs tested had significantly lower luciferase activity compared with the vector alone (P < 0.01). C, Activity of strain-specific luciferase-3′ UTR constructs in transiently transfected B6 or C3H stromal cells. Luciferase activity is expressed as a percentage of β-gal activity (mean ±
sem
).
Fig. 2.
Igf1 splice variants in osteoblasts and identification of 3′ ends for short Igf1 mRNAs. A, Diagram of alternative splicing patterns for mouse Igf1. Polyadenylation sites in exon 6 are indicated by arrows, and UTRs are indicated in gray. B, 3′ RACE design. C, Short Igf1 mRNA ends identified. The most common site of polyA addition is denoted by the asterisk, and less common sites are denoted by the caret. The miR-1/206 binding site is underlined, and a miR-365 site is double underlined. The polyA signal is in italic.
Fig. 3.
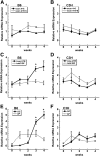
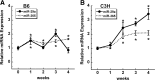
Evaluation of Igf1 splice variants and 3′ UTR length between C3H and B6 cells during osteoblast differentiation in vitro. B6 and C3H stromal cells were grown to confluence (0) and then cultured for up to 4 wk after confluence in osteoblast differentiation medium. RNA abundance was quantified by qRT-PCR and expressed relative to 18S rRNA. *, Significantly different from confluence, P < 0.05. A and B, Splice variants. C and D, Total Igf1 mRNA (amplified by an exon 4 primer set) compared with mRNA containing the long exon 6 3′ UTR. E and F, Alkaline phosphatase (AP) and osteocalcin (OC) mRNA levels, documenting the differentiation state of the cultures.
Fig. 4.
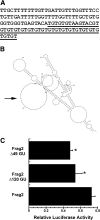
GU-rich motif in long Igf1 3′ UTR. A, Sequence of GU-rich motif (B6 3′ UTR bases 4023–4143). Region deleted for the Δ49 GU construct is underlined. B, Potential secondary structure (sFOLD) of GU18 element, in the context of the 3275–5774 fragment 2 (Frag2). The potential secondary structure of this region was similar in the context of the 1–6292 fragment 4 and was not altered by strain-specific polymorphisms. C, Deletion of the entire GU-rich motif (Δ120 GU), or the region containing GU18 (Δ49 GU), decreases 3′ UTR-regulated luciferase activity, in the context of fragment 2 (3275–5779). *, Significantly different from intact fragment 2, P < 0.05.
Fig. 5.
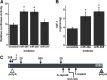
miR-29a and miR-365 target IGF-I. A, Activity of luciferase-1–6292 3′ UTR construct (fragment 4) in MC3T3 cells, transiently cotransfected with 50 n
m
miRNA inhibitor or scrambled control. *, Significantly different from scrambled control, P < 0.05. B, Quantity of IGF-I in conditioned medium from MC3T3 cells, transiently transfected with 50 n
m
miRNA inhibitor or scrambled control. *, Significantly different from scrambled control, P < 0.05. IGF-I was quantified by RIA and expressed as ng/ml·mg cell layer protein. C, Schematic representation of the Igf1 3′ UTR from exon 6, with the relative location of sequence motifs and binding sties for miR-365, miR-1, and miR-29.
Fig. 6.
miR-29a and miR-365 increase during osteoblastic differentiation in vitro. B6 (A) and C3H (B) stromal cells were grown to confluence (0) and then cultured for up to 4 wk after confluence in osteoblast differentiation medium. miRNA abundance was quantified by qRT-PCR and expressed relative to 5S rRNA (for miR-29a) or Sno202 (for miR-365). *, Significantly different from confluence, P < 0.05.
Similar articles
- Nocturnin suppresses igf1 expression in bone by targeting the 3' untranslated region of igf1 mRNA.
Kawai M, Delany AM, Green CB, Adamo ML, Rosen CJ. Kawai M, et al. Endocrinology. 2010 Oct;151(10):4861-70. doi: 10.1210/en.2010-0407. Epub 2010 Aug 4. Endocrinology. 2010. PMID: 20685873 Free PMC article. - Congenic mice with low serum IGF-I have increased body fat, reduced bone mineral density, and an altered osteoblast differentiation program.
Rosen CJ, Ackert-Bicknell CL, Adamo ML, Shultz KL, Rubin J, Donahue LR, Horton LG, Delahunty KM, Beamer WG, Sipos J, Clemmons D, Nelson T, Bouxsein ML, Horowitz M. Rosen CJ, et al. Bone. 2004 Nov;35(5):1046-58. doi: 10.1016/j.bone.2004.07.008. Bone. 2004. PMID: 15542029 - Antisense oligonucleotide-mediated redirection of Igf1 alternative polyadenylation.
Yavas A, van Putten M, Ariyurek Y, Aartsma-Rus A. Yavas A, et al. Neuromuscul Disord. 2025 Jun;51:105394. doi: 10.1016/j.nmd.2025.105394. Epub 2025 May 24. Neuromuscul Disord. 2025. PMID: 40450413 - The complexity of the IGF1 gene splicing, posttranslational modification and bioactivity.
Philippou A, Maridaki M, Pneumaticos S, Koutsilieris M. Philippou A, et al. Mol Med. 2014 May 7;20(1):202-14. doi: 10.2119/molmed.2014.00011. Mol Med. 2014. PMID: 24637928 Free PMC article. Review. - Role of Alternatively Spliced Messenger RNA (mRNA) Isoforms of the Insulin-Like Growth Factor 1 (IGF1) in Selected Human Tumors.
Kasprzak A, Szaflarski W. Kasprzak A, et al. Int J Mol Sci. 2020 Sep 23;21(19):6995. doi: 10.3390/ijms21196995. Int J Mol Sci. 2020. PMID: 32977489 Free PMC article. Review.
Cited by
- Anabolic effects of IGF-1 signaling on the skeleton.
Tahimic CG, Wang Y, Bikle DD. Tahimic CG, et al. Front Endocrinol (Lausanne). 2013 Feb 4;4:6. doi: 10.3389/fendo.2013.00006. eCollection 2013. Front Endocrinol (Lausanne). 2013. PMID: 23382729 Free PMC article. - Post-transcriptional regulation in osteoblasts using localized delivery of miR-29a inhibitor from nanofibers to enhance extracellular matrix deposition.
James EN, Delany AM, Nair LS. James EN, et al. Acta Biomater. 2014 Aug;10(8):3571-80. doi: 10.1016/j.actbio.2014.04.026. Epub 2014 May 9. Acta Biomater. 2014. PMID: 24816265 Free PMC article. - Regulatory Role of MicroRNAs in Muscle Atrophy during Exercise Intervention.
Zhang S, Chen N. Zhang S, et al. Int J Mol Sci. 2018 Jan 30;19(2):405. doi: 10.3390/ijms19020405. Int J Mol Sci. 2018. PMID: 29385720 Free PMC article. Review. - Pro-apoptotic effects of micro-ribonucleic acid-365 on retinal neurons by targeting insulin-like growth factor-1 in diabetic rats: An in vivo and in vitro study.
Zheng K, Wang N, Shen Y, Zhang Z, Gu Q, Xu X, Qin Q, Liu Y. Zheng K, et al. J Diabetes Investig. 2018 Sep;9(5):1041-1051. doi: 10.1111/jdi.12815. Epub 2018 May 14. J Diabetes Investig. 2018. PMID: 29427460 Free PMC article. - Mega roles of microRNAs in regulation of skeletal muscle health and disease.
Sharma M, Juvvuna PK, Kukreti H, McFarlane C. Sharma M, et al. Front Physiol. 2014 Jun 26;5:239. doi: 10.3389/fphys.2014.00239. eCollection 2014. Front Physiol. 2014. PMID: 25018733 Free PMC article. Review.
References
- Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, Pohl A, Malladi VS, Li CH, Learned K, Kirkup V, Hsu F, Harte RA, Guruvadoo L, Goldman M, Giardine BM, Fujita PA, Diekhans M, Cline MS, Clawson H, Barber GP, Haussler D, James Kent W. 2012. The UCSC genome browser database: extensions and updates 2011. Nucleic Acids Res 40:D918–D923 - PMC - PubMed
- Barton ER. 2006. The ABCs of IGF-1 isoforms: impact on muscle hypertrophy and implications for repair. Appl Physiol Nutr Metab 31:791–797 - PubMed
- Foyt HL, LeRoith D, Roberts CT., Jr 1991. Differential association of insulin-like growth factor I mRNA variants with polysomes in vivo. J Biol Chem 266:7300–7305 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR045433/AR/NIAMS NIH HHS/United States
- R56 AR044877/AR/NIAMS NIH HHS/United States
- AR44877/AR/NIAMS NIH HHS/United States
- R01 AR044877/AR/NIAMS NIH HHS/United States
- AR45433/AR/NIAMS NIH HHS/United States
- R29 AR044877/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous