Word decoding of protein amino Acid sequences with availability analysis: a linguistic approach - PubMed (original) (raw)
Word decoding of protein amino Acid sequences with availability analysis: a linguistic approach
Kenta Motomura et al. PLoS One. 2012.
Abstract
The amino acid sequences of proteins determine their three-dimensional structures and functions. However, how sequence information is related to structures and functions is still enigmatic. In this study, we show that at least a part of the sequence information can be extracted by treating amino acid sequences of proteins as a collection of English words, based on a working hypothesis that amino acid sequences of proteins are composed of short constituent amino acid sequences (SCSs) or "words". We first confirmed that the English language highly likely follows Zipf's law, a special case of power law. We found that the rank-frequency plot of SCSs in proteins exhibits a similar distribution when low-rank tails are excluded. In comparison with natural English and "compressed" English without spaces between words, amino acid sequences of proteins show larger linear ranges and smaller exponents with heavier low-rank tails, demonstrating that the SCS distribution in proteins is largely scale-free. A distribution pattern of SCSs in proteins is similar among species, but species-specific features are also present. Based on the availability scores of SCSs, we found that sequence motifs are enriched in high-availability sites (i.e., "key words") and vice versa. In fact, the highest availability peak within a given protein sequence often directly corresponds to a sequence motif. The amino acid composition of high-availability sites within motifs is different from that of entire motifs and all protein sequences, suggesting the possible functional importance of specific SCSs and their compositional amino acids within motifs. We anticipate that our availability-based word decoding approach is complementary to sequence alignment approaches in predicting functionally important sites of unknown proteins from their amino acid sequences.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
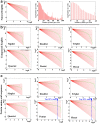
Figure 1. Distributions of English letters and protein amino acid sequences.
The relationships between rank (R) and number (frequency) of words (N) are shown in the log-log plots. The background red lines indicate Y = A – bX, where b varies with an interval of 0.05. The thick red line indicates the best-fit least squares line. The green dots and lines indicate the observed results. (a) Natural English, following Zipf's law (left), and distribution of English word lengths (middle and right). The word lengths peak at 3. (b) Compressed English. (c) Protein amino acid sequences.
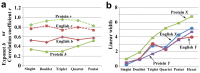
Figure 2. The exponent and linear widths of compressed English and proteins in the rank-frequency plot.
(a) The exponent b and correlation coefficient r in compressed English and proteins. (b) The linear width in compressed English and proteins both in _X_- and _Y_-axes.
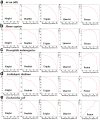
Figure 3. Relationship between remaining top rank sample (%) and discriminant R value.
(a) nr-aa (all). (b) H. sapiens. (c) D. melanogaster. (d) A. thaliana. (e) E. coli.
Figure 4. The rank-frequency relationship of proteins in four model organisms.
Boundaries at the top 50% rank samples were indicated by blue dotted line. (a) H. sapiens. (b) D. melanogaster. (c) A. thaliana. (d) E. coli.
Figure 5. The exponents and linear widths of proteins from four model organisms in the rank-frequency plots.
(a) The exponent b. (b) The correlation coefficient r. (c) The linear width of _X_-axis. (d) Linear width of _Y_-axis.
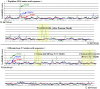
Figure 6. The availability plots of two examples of amino acid sequences of proteins, peptidase M10 (top) and ribonuclease T2 (bottom).
_X_-and _Y_-axes indicate protein amino acid sequence and availability score, respectively.
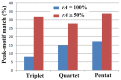
Figure 7. The percentages of correspondence between sequence motifs and SCSs (i.e., triplets, quartets, and pentats).
The blue bars indicate rA = 100%, whereas the red bars indicate _rA_≥50%.
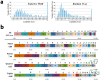
Figure 8. The characterization of high-availability sites within motifs.
(a) The distribution of the pentat availability scores within motifs and random fragments. (b) The rank order analysis of the amino acid composition of full sequences, motifs, and high-availability areas of pentats, quartets, and triplets within motifs. The rank order distance (ROD) scores are shown beside the horizontal bars. The numbers at the top of each amino acid indicate the rank difference from the full sequences.
Similar articles
- A frequency-based linguistic approach to protein decoding and design: Simple concepts, diverse applications, and the SCS Package.
Motomura K, Nakamura M, Otaki JM. Motomura K, et al. Comput Struct Biotechnol J. 2013 Mar 29;5:e201302010. doi: 10.5936/csbj.201302010. eCollection 2013. Comput Struct Biotechnol J. 2013. PMID: 24688703 Free PMC article. Review. - Potential implications of availability of short amino acid sequences in proteins: an old and new approach to protein decoding and design.
Otaki JM, Gotoh T, Yamamoto H. Otaki JM, et al. Biotechnol Annu Rev. 2008;14:109-41. doi: 10.1016/S1387-2656(08)00004-5. Biotechnol Annu Rev. 2008. PMID: 18606361 Review. - Improving protein secondary structure prediction based on short subsequences with local structure similarity.
Lin HN, Sung TY, Ho SY, Hsu WL. Lin HN, et al. BMC Genomics. 2010 Dec 2;11 Suppl 4(Suppl 4):S4. doi: 10.1186/1471-2164-11-S4-S4. BMC Genomics. 2010. PMID: 21143813 Free PMC article. - Zipf's word frequency law in natural language: a critical review and future directions.
Piantadosi ST. Piantadosi ST. Psychon Bull Rev. 2014 Oct;21(5):1112-30. doi: 10.3758/s13423-014-0585-6. Psychon Bull Rev. 2014. PMID: 24664880 Free PMC article. Review. - Functional proteomics with biolinguistic methods. n-grams deliver sensitive portrayals of gene similarity.
Singh GB, Singh H. Singh GB, et al. IEEE Eng Med Biol Mag. 2005 May-Jun;24(3):73-80. doi: 10.1109/memb.2005.1436463. IEEE Eng Med Biol Mag. 2005. PMID: 15971844 No abstract available.
Cited by
- The Compressed Vocabulary of Microbial Life.
Caetano-Anollés G. Caetano-Anollés G. Front Microbiol. 2021 Jul 7;12:655990. doi: 10.3389/fmicb.2021.655990. eCollection 2021. Front Microbiol. 2021. PMID: 34305827 Free PMC article. - Computer-Aided Design of Antimicrobial Peptides: Are We Generating Effective Drug Candidates?
Cardoso MH, Orozco RQ, Rezende SB, Rodrigues G, Oshiro KGN, Cândido ES, Franco OL. Cardoso MH, et al. Front Microbiol. 2020 Jan 22;10:3097. doi: 10.3389/fmicb.2019.03097. eCollection 2019. Front Microbiol. 2020. PMID: 32038544 Free PMC article. Review. - The estimation of probability distribution for factor variables with many categorical values.
Lee M, Kang YS, Seok J. Lee M, et al. PLoS One. 2018 Aug 24;13(8):e0202547. doi: 10.1371/journal.pone.0202547. eCollection 2018. PLoS One. 2018. PMID: 30142178 Free PMC article. - Quantiprot - a Python package for quantitative analysis of protein sequences.
Konopka BM, Marciniak M, Dyrka W. Konopka BM, et al. BMC Bioinformatics. 2017 Jul 17;18(1):339. doi: 10.1186/s12859-017-1751-4. BMC Bioinformatics. 2017. PMID: 28716000 Free PMC article.
References
- Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181: 223–230. - PubMed
- Searls DB (2002) The language of genes. Nature 420: 211–217. - PubMed
- Searls DB (1997) Linguistic approaches to biological sequences. Comput Appl Biosci 13: 333–344. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This work was supported by funds from University of the Ryukyus, Okinawa, Japan. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources