Heterogeneous expression of the core circadian clock proteins among neuronal cell types in mouse retina - PubMed (original) (raw)
Heterogeneous expression of the core circadian clock proteins among neuronal cell types in mouse retina
Xiaoqin Liu et al. PLoS One. 2012.
Abstract
Circadian rhythms in metabolism, physiology, and behavior originate from cell-autonomous circadian clocks located in many organs and structures throughout the body and that share a common molecular mechanism based on the clock genes and their protein products. In the mammalian neural retina, despite evidence supporting the presence of several circadian clocks regulating many facets of retinal physiology and function, the exact cellular location and genetic signature of the retinal clock cells remain largely unknown. Here we examined the expression of the core circadian clock proteins CLOCK, BMAL1, NPAS2, PERIOD 1(PER1), PERIOD 2 (PER2), and CRYPTOCHROME2 (CRY2) in identified neurons of the mouse retina during daily and circadian cycles. We found concurrent clock protein expression in most retinal neurons, including cone photoreceptors, dopaminergic amacrine cells, and melanopsin-expressing intrinsically photosensitive ganglion cells. Remarkably, diurnal and circadian rhythms of expression of all clock proteins were observed in the cones whereas only CRY2 expression was found to be rhythmic in the dopaminergic amacrine cells. Only a low level of expression of the clock proteins was detected in the rods at any time of the daily or circadian cycle. Our observations provide evidence that cones and not rods are cell-autonomous circadian clocks and reveal an important disparity in the expression of the core clock components among neuronal cell types. We propose that the overall temporal architecture of the mammalian retina does not result from the synchronous activity of pervasive identical clocks but rather reflects the cellular and regional heterogeneity in clock function within retinal tissue.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
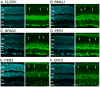
Figure 1. Distribution of six core mammalian circadian clock proteins in the mouse retina.
Typical examples of vertical sections of C57Bl/6 mouse retinas collected in the middle of the day during a LD cycle (ZT02-06) are illustrated for CLOCK (A), BMAL1 (B), NPAS2 (C), PER1 (D), PER2 (E), and CRY2 (F). Expression of each clock protein is clearly prominent in the inner nuclear layer (INL) and most cells in the ganglion cell layer (GCL). However, expression is also detected in few cells in the outer nuclear layer (ONL) (vertical arrows). These cells were later identified as cones. OPL: outer plexiform layer; IPL: inner plexiform layer. Optical sections 3×0.4 µm. Bar is 10 µm.
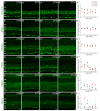
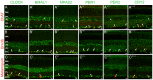
Figure 2. Expression of the six core mammalian circadian clock proteins CLOCK, BMAL1, NPAS2, PER1, PER2 and CRY2 in mouse retina during LD and DD cycles.
Typical examples of vertical sections of C57Bl/6 mouse retinas collected in the middle of the day (ZT06; A–F) or subjective day (CT06; A”–F”) or middle of the night (ZT18; A’–F’) or subjective night (CT18; A”’–F”’) are illustrated for CLOCK (A–A”’), BMAL1 (B–B”’), NPAS2 (C–C”’), PER1 (D–D”’), PER2 (E–E”’), and CRY2 (F–F”’). Optical sections 3×0.4 µm. Bar is 10 µm. Temporal expression profiles of CLOCK, BMAL1, NPAS2, PER1, PER2, and CRY2 in mouse retinal sections under LD and DD conditions (A””–F””). Mouse retinas were collected every 4 h during a LD cycle or a DD cycle, and clock protein expression was quantified by measuring the mean fluorescence per section within a squared area whose upper and lower boundaries were the outer and inner limiting membranes, respectively (see Materials and Methods for details). COSINOR regression was performed only when variations in clock protein expression with time of day were detected (one-way ANOVA; P<0.05). Note that only PER1 expression is significantly rhythmic under either LD or DD conditions (D””). Each data point represents the mean fluorescence/section +/− SEM of 5/6 animals (5 pictures from 3–4 sections/animal). ONL: outer nuclear layer; OPL: outer plexiform layer; INL: inner nuclear layer; IPL: inner plexiform layer; GCL: ganglion cell layer.
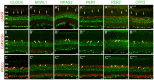
Figure 3. Mammalian core circadian clock protein expression in cones, horizontal cells and rod bipolar cells of the mouse retina.
Typical examples of vertical sections of mouse retinas collected between ZT02 and ZT06 and double labeled for one of the following clock proteins: CLOCK (A – C), BMAL1 (A ’– C ’), NPAS2 (A ”– C ”), PER1 (A ’”– C ’”), PER2 (A ””– C ””), and CRY2 (A ’””– C ’””) and one of the following protein markers: cARR (cones; A), CALD28 (horizontal cells; B), and PKCα (rod bipolar cells; C). (see Table 1 for details about the antibodies). Note that in the outer nuclear layer (ONL) clock protein expression was evident in cones but was weak or absent in rods. Some double-labeled retinal neurons are shown (arrows). OPL: outer plexiform layer; INL: inner nuclear layer; IPL: inner plexiform layer. Optical sections 3×0.4 µm. Bar is 10 µm.
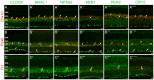
Figure 4. Mammalian core circadian clock protein expression in bipolar, amacrine and ganglion cells of the mouse retina.
Typical examples of vertical sections of mouse retinas collected between ZT02 and ZT06 and double labeled for one of the following clock proteins: CLOCK (A–C), BMAL1 (A’–C’), NPAS2 (A”–C”), PER1 (A’”–C’”), PER2 (A””–C””), and CRY2 (A’””–C’””) and one of the following protein markers: Chx10 (bipolar cells; A), Pax6 (most amacrine cells and ganglion cells; B), and TH (dopaminergic amacrine cells; C) (see Table 1 for details about the antibodies). The analysis was restricted to type-1 catecholamine amacrine cells that express high levels of TH. Some double-labeled retinal neurons are shown (arrows). Abbreviations and bar as in Fig. 3.
Figure 5. Mammalian core circadian clock protein expression in amacrine and ganglion cells of the mouse retina.
Typical examples of vertical sections of mouse retinas collected between ZT02 and ZT06 and double labeled for one of the following clock proteins: CLOCK (A–C), BMAL1 (A’–C’), NPAS2 (A”–C”), PER1 (A’”–C’”), PER2 (A””–C””), and CRY2 (A’””–C’””) and one of the following protein markers: ChAT (starburst amacrine cells; A), Brn3b (most ganglion cells; B), and eGFP (ipRGCs; C) (see Table 1 for details about the antibodies). The concurrent expression of CLOCK, BMAL1, NPAS2, PER1, PER2, and CRY2 was found in all identified neurons. Note also that all the clock proteins are expressed in the ON starburst amacrine cells whose cell body is located in the ganglion cell layer (GCL), indicating that both GCs and displaced amacrine cells in the GCL express the core components of the mammalian clock. Clock protein expression in ipRGCs was confirmed with the AB-N38 antibody (data not illustrated), although this antibody only labeled M1 and M2 ipRGC subtypes. Some double-labeled retinal neurons are shown (arrows). Abbreviations and bar as in Fig. 3.
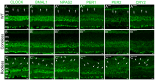
Figure 6. Core circadian clock protein expression in wild-type, coneless and rodeless retinas.
Typical examples of vertical sections of wild-type C57Bl/6J (A–A’””), coneless (B–B’””), and rodeless (C–C’””) retinas immunolabeled for each of the following core clock proteins: CLOCK (A–C), BMAL1 (A’–C’), NPAS2 (A”–C”), PER1 (A’”–C’”), PER2 (A””–C””), and CRY2 (A’””–C’””). Retinal tissue was collected around ZT09. For a given clock protein antibody, confocal settings were adjusted on the brightest picture and the 2 other sections were taken at the same settings. Note that clock protein expression in the outer nuclear layer (ONL) is detected in a few cells in the wild-type retina (vertical arrows) and in most cells in the rodless retina (oblique arrows), but is very weak in the coneless retina. OPL: outer plexiform layer; INL: inner nuclear layer; IPL: inner plexiform layer; GCL: ganglion cell layer. Optical sections 3×0.4 µm. Bar is 10 µm.
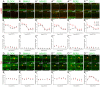
Figure 7. Circadian clock core component expression in mouse rod and cone photoreceptors and dopaminergic amacrine cells under LD and DD conditions.
Typical examples of clock protein immunostaining in cones (A – A ””’) and dopaminergic cells (D – D ’””) obtained from retinas collected in the middle of the day (ZT06) or subjective day (CT06) or middle of the night (ZT18) or subjective night (CT18) are illustrated for CLOCK (A , D), BMAL1 (A ’, D ’), NPAS2 (A ”, D ”), PER1 (A ’”, D ’”), PER2 (A ””, D ””), and CRY2 (A ’””, D ’””). COSINOR regression analysis (cosine curves) was performed only for clock protein levels displaying significant temporal variation (as determined by one-way ANOVA; P<0.05). The results from the COSINOR analysis are shown in Table 4. Note that the expression of all six clock proteins is rhythmic under LD and DD conditions in cones (B – B ’””) but arrhythmic under the same conditions in dopaminergic amacrine cells, except for CRY which is rhythmic under both LD and DD conditions (E – E ’””). Clock protein expression in rods remained low at any time point under both LD and DD conditions (C – C ’””). Each data point represents the mean fluorescence/identified neuron +/− SEM of 5/6 animals (5 pictures from 3–4 sections/animal).
Similar articles
- Divergent roles of clock genes in retinal and suprachiasmatic nucleus circadian oscillators.
Ruan GX, Gamble KL, Risner ML, Young LA, McMahon DG. Ruan GX, et al. PLoS One. 2012;7(6):e38985. doi: 10.1371/journal.pone.0038985. Epub 2012 Jun 11. PLoS One. 2012. PMID: 22701739 Free PMC article. - Quantification of interactions among circadian clock proteins via surface plasmon resonance.
Kepsutlu B, Kizilel R, Kizilel S. Kepsutlu B, et al. J Mol Recognit. 2014 Jul;27(7):458-69. doi: 10.1002/jmr.2367. J Mol Recognit. 2014. PMID: 24895278 - Daily rhythmic expression patterns of clock1a, bmal1, and per1 genes in retina and hypothalamus of the rainbow trout, Oncorhynchus mykiss.
Patiño MA, Rodríguez-Illamola A, Conde-Sieira M, Soengas JL, Míguez JM. Patiño MA, et al. Chronobiol Int. 2011 May;28(5):381-9. doi: 10.3109/07420528.2011.566398. Chronobiol Int. 2011. PMID: 21721853 - [Molecular mechanisms of circadian clock functioning].
Karbovskyĭ LL, Minchenko DO, Garmash IaA, Minchenko OG. Karbovskyĭ LL, et al. Ukr Biokhim Zh (1999). 2011 May-Jun;83(3):5-24. Ukr Biokhim Zh (1999). 2011. PMID: 21888051 Review. Ukrainian. - Influence of the extracellular matrix on cell-intrinsic circadian clocks.
Streuli CH, Meng QJ. Streuli CH, et al. J Cell Sci. 2019 Feb 1;132(3):jcs207498. doi: 10.1242/jcs.207498. J Cell Sci. 2019. PMID: 30709969 Review.
Cited by
- Glial Bmal1 role in mammalian retina daily changes.
Riccitelli S, Boi F, Lonardoni D, Giantomasi L, Barca-Mayo O, De Pietri Tonelli D, Bisti S, Di Marco S, Berdondini L. Riccitelli S, et al. Sci Rep. 2022 Dec 13;12(1):21561. doi: 10.1038/s41598-022-25783-1. Sci Rep. 2022. PMID: 36513717 Free PMC article. - Circadian regulation in the retina: From molecules to network.
Ko GY. Ko GY. Eur J Neurosci. 2020 Jan;51(1):194-216. doi: 10.1111/ejn.14185. Epub 2018 Oct 24. Eur J Neurosci. 2020. PMID: 30270466 Free PMC article. Review. - Neuronal Bmal1 regulates retinal angiogenesis and neovascularization in mice.
Jidigam VK, Sawant OB, Fuller RD, Wilcots K, Singh R, Lang RA, Rao S. Jidigam VK, et al. Commun Biol. 2022 Aug 6;5(1):792. doi: 10.1038/s42003-022-03774-2. Commun Biol. 2022. PMID: 35933488 Free PMC article. - Pre-mRNA Processing Factors and Retinitis Pigmentosa: RNA Splicing and Beyond.
Yang C, Georgiou M, Atkinson R, Collin J, Al-Aama J, Nagaraja-Grellscheid S, Johnson C, Ali R, Armstrong L, Mozaffari-Jovin S, Lako M. Yang C, et al. Front Cell Dev Biol. 2021 Jul 28;9:700276. doi: 10.3389/fcell.2021.700276. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34395430 Free PMC article. Review. - Rat retina shows robust circadian expression of clock and clock output genes in explant culture.
Buonfiglio DC, Malan A, Sandu C, Jaeger C, Cipolla-Neto J, Hicks D, Felder-Schmittbuhl MP. Buonfiglio DC, et al. Mol Vis. 2014 Jun 2;20:742-52. eCollection 2014. Mol Vis. 2014. PMID: 24940028 Free PMC article.
References
- Dunlap JC, Loros JJ, DeCoursey PJ (2003) Chronobiology. Biological Timekeeping. Sunderland, MA: Sinauer Associates, Inc. 406 p.
- Barlow R (2001) Circadian and efferent modulation of visual sensitivity. Prog Brain Res 131: 487–503. - PubMed
- Green CB, Besharse JC (2004) Retinal circadian clocks and control of retinal physiology. J Biol Rhythms 19: 91–102. - PubMed
- Iuvone PM, Tosini G, Pozdeyev N, Haque R, Klein DC, et al. (2005) Circadian clocks, clock-controlled genes and melatonin biosynthesis in the retina. Prog Retin Eye Res 24: 433–456. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases