Mechanisms of antibiotic resistance in Burkholderia pseudomallei: implications for treatment of melioidosis - PubMed (original) (raw)
Review
Mechanisms of antibiotic resistance in Burkholderia pseudomallei: implications for treatment of melioidosis
Herbert P Schweizer. Future Microbiol. 2012 Dec.
Abstract
Burkholderia pseudomallei is the etiologic agent of melioidosis. This multifaceted disease is difficult to treat, resulting in high morbidity and mortality. Treatment of B. pseudomallei infections is lengthy and necessitates an intensive phase (parenteral ceftazidime, amoxicillin-clavulanic acid or meropenem) and an eradication phase (oral trimethoprim-sulfamethoxazole). The main resistance mechanisms affecting these antibiotics include enzymatic inactivation, target deletion and efflux from the cell, and are mediated by chromosomally encoded genes. Overproduction and mutations in the class A PenA β-lactamase cause ceftazidime and amoxicillin-clavulanic acid resistance. Deletion of the penicillin binding protein 3 results in ceftazidime resistance. BpeEF-OprC efflux pump expression causes trimethoprim and trimethoprim-sulfamethoxazole resistance. Although resistance is still relatively rare, therapeutic efficacies may be compromised by resistance emergence due to increased use of antibiotics in endemic regions. Novel agents and therapeutic strategies are being tested and, in some instances, show promise as anti-B. pseudomallei infectives.
Conflict of interest statement
Financial & competing interests disclosure
The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Figures
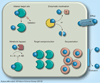
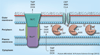
Figure 1. Bacterial antibiotic resistance mechanisms
Bacterial antimicrobial resistance mechanisms are multifaceted, ranging from: exclusion of drug molecules (blue spheres) from the bacterial cell by physicochemical constraints (e.g., porins or lipopolysaccharide); efflux from the cell via active transport mechanisms; enzymatic inactivation, either in the form of substrate cleavage or chemical modification (M = acetylation, adenylation or phosphorylation); target site alteration or, rarely, target deletion; metabolic bypass by substitution of a susceptible enzyme or pathway with a resistant enzyme or pathway; target overproduction by either increased transcription or gene multiplication; and drug sequestration by specific binding proteins akin to immunity proteins. Details about individual resistance mechanisms and specific examples are provided in the text. Bacteria often employ different resistance mechanisms that act synergistically, for instance, efflux and exclusion, to achieve high-level resistance [26].
Figure 2. Burkholderia pseudomallei PenA and locations of mutations leading to clinically significant antibiotic resistance
Positions of conserved regions and mutations are numbered according to the Ambler scheme [69]. Susceptibilities to clinically significant β-lactam antibiotics range from 1.5 to 3 µg/ml (wild-type) [33,34,36,64], ≥256 µg/ml (C69Y) [33,34,64] and 24 to 64 µg/ml (P167S) [36,64] for ceftazidime, and 3 to 8 µg/ml (wild-type) and 16 to 32 µg/ml (P72F) for amoxicillin–clavulanic acid [64,67]. By comparison, the meropenem MICs in all isolates are not significantly affected and range from 0.75 to 1.5 µg/ml [36,64]. Current susceptibility breakpoints are ≤8 µg/ml for ceftazidime, ≤8 µg/ml for amoxicillin and ≤4 µg/ml for clavulanic acid.
Figure 3. Summary of Burkholderia pseudomallei resistance mechanisms compromising therapy with clinically significant antibiotics
Enzymatic inactivation and efflux from the cell are mechanisms that compromise the use of antibiotics employed in intensive and eradication phase therapy. The CEF and AMX targets are located in the periplasm. There is experimental evidence that CEF and other β-lactams permeate the outer membrane through porins. Periplasmic β-lactams, such as AMX, are (A) inactivated by the wild-type PenA class A β-lactamase, (B) unless its activity is inhibited by CLA. Although wild-type PenA hydrolyzes CEF to some extent, it does not confer clinically significant resistance to CEF. (C) Some mutant PenA (PenAmt) derivatives catalyze CEF hydrolysis and others (not illustrated) are refractory to CLA inhibition. Antibiotics such as DOX, TMP and TMP–SMX have cytoplasmic targets and are extruded to the extracellular milieu by the multicomponent BpeEF–OprC resistance nodulation and cell division efflux pump. As resistance nodulation and cell division pumps extrude substrates acquired from the periplasmic space, the listed antibiotics are most likely extruded either during their transit into the cell or after extrusion to the periplasm from the cytoplasmic space via an unknown mechanism. AMX: Amoxicillin; CEF: Ceftazidime; CLA: Clavulanic acid; DOX: Doxycycline; SMX: Sulfamethoxazole; TMP: Trimethoprim.
Similar articles
- Mechanisms of Resistance to Folate Pathway Inhibitors in Burkholderia pseudomallei: Deviation from the Norm.
Podnecky NL, Rhodes KA, Mima T, Drew HR, Chirakul S, Wuthiekanun V, Schupp JM, Sarovich DS, Currie BJ, Keim P, Schweizer HP. Podnecky NL, et al. mBio. 2017 Sep 5;8(5):e01357-17. doi: 10.1128/mBio.01357-17. mBio. 2017. PMID: 28874476 Free PMC article. - Antibiotic Resistance Markers in Burkholderia pseudomallei Strain Bp1651 Identified by Genome Sequence Analysis.
Bugrysheva JV, Sue D, Gee JE, Elrod MG, Hoffmaster AR, Randall LB, Chirakul S, Tuanyok A, Schweizer HP, Weigel LM. Bugrysheva JV, et al. Antimicrob Agents Chemother. 2017 May 24;61(6):e00010-17. doi: 10.1128/AAC.00010-17. Print 2017 Jun. Antimicrob Agents Chemother. 2017. PMID: 28396541 Free PMC article. - Transcriptional and post-transcriptional regulation of PenA β-lactamase in acquired Burkholderia pseudomallei β-lactam resistance.
Chirakul S, Norris MH, Pagdepanichkit S, Somprasong N, Randall LB, Shirley JF, Borlee BR, Lomovskaya O, Tuanyok A, Schweizer HP. Chirakul S, et al. Sci Rep. 2018 Jul 13;8(1):10652. doi: 10.1038/s41598-018-28843-7. Sci Rep. 2018. PMID: 30006637 Free PMC article. - Treatment and prophylaxis of melioidosis.
Dance D. Dance D. Int J Antimicrob Agents. 2014 Apr;43(4):310-8. doi: 10.1016/j.ijantimicag.2014.01.005. Epub 2014 Feb 3. Int J Antimicrob Agents. 2014. PMID: 24613038 Free PMC article. Review. - Antibiotic resistance in Burkholderia species.
Rhodes KA, Schweizer HP. Rhodes KA, et al. Drug Resist Updat. 2016 Sep;28:82-90. doi: 10.1016/j.drup.2016.07.003. Epub 2016 Jul 30. Drug Resist Updat. 2016. PMID: 27620956 Free PMC article. Review.
Cited by
- Protein expression, purification, crystallization and crystallographic studies of BPSL0741 from Burkholderia pseudomallei.
Fadhar NF, Nyanasegran PK, Firdaus-Raih M, Nathan S, Jonet MA, Ng CL. Fadhar NF, et al. Acta Crystallogr F Struct Biol Commun. 2024 Oct 1;80(Pt 10):263-268. doi: 10.1107/S2053230X24008197. Epub 2024 Sep 11. Acta Crystallogr F Struct Biol Commun. 2024. PMID: 39259140 - Structural and functional diversity of Resistance-Nodulation-Division (RND) efflux pump transporters with implications for antimicrobial resistance.
Kavanaugh LG, Dey D, Shafer WM, Conn GL. Kavanaugh LG, et al. Microbiol Mol Biol Rev. 2024 Sep 26;88(3):e0008923. doi: 10.1128/mmbr.00089-23. Epub 2024 Sep 5. Microbiol Mol Biol Rev. 2024. PMID: 39235227 Review. - A molecular epidemiological analysis of Burkholderia pseudomallei in southern Thailand.
Kaewrakmuk J, Chusri S, Khrongsee P, Kawila S, Saechan V, Leesahud N, Chiewchanyont B, Thananchai H, Duangsonk K, Tuanyok A. Kaewrakmuk J, et al. PLoS Negl Trop Dis. 2024 Aug 22;18(8):e0012444. doi: 10.1371/journal.pntd.0012444. eCollection 2024 Aug. PLoS Negl Trop Dis. 2024. PMID: 39173078 Free PMC article. - Complete genome sequences of paired isogenic Burkholderia pseudomallei isolated from a Thai pediatric patient with a urinary tract infection.
Chirakul S, Suchartlikitwong P, Saninjuk K, Faksri K, Patarakul K, Jenjaroenpun P, Wongsurawat T. Chirakul S, et al. Microbiol Resour Announc. 2024 Aug 13;13(8):e0047024. doi: 10.1128/mra.00470-24. Epub 2024 Jul 8. Microbiol Resour Announc. 2024. PMID: 38975774 Free PMC article. - Epidemiology and genetic diversity of Burkholderia pseudomallei from Riau Province, Indonesia.
Anggraini D, Siregar FM, Rosdiana D, Kemal RA, Yovi I, Triani ZD, Jasmin N, Dwijelita N, Webb JR, Mayo M, Kaestli M, Currie BJ. Anggraini D, et al. PLoS Negl Trop Dis. 2024 May 28;18(5):e0012195. doi: 10.1371/journal.pntd.0012195. eCollection 2024 May. PLoS Negl Trop Dis. 2024. PMID: 38805481 Free PMC article.
References
- Ashdown LR, Duffy VA, Douglas RA. Melioidosis. Med. J. Aust. 1980;1:314–316. - PubMed
- White NJ. Melioidosis. Lancet. 2003;361(9370):1715–1722. - PubMed
- Leelarasamee A. Recent development in melioidosis. Curr. Opin. Infect. Dis. 2004;17:131–136. - PubMed
- Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 2005;18(2):383–416. ▪▪ Excellent review on the various aspects of melioidosis.
- Wiersinga WJ, Van Der Poll T, White NJ, Day NP, Peacock SJ. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 2006;4:272–282. ▪▪ Excellent review of Burkholderia pseudomallei and melioidosis.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical