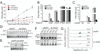Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway - PubMed (original) (raw)
Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway
Lijun Sun et al. Science. 2013.
Abstract
The presence of DNA in the cytoplasm of mammalian cells is a danger signal that triggers host immune responses such as the production of type I interferons. Cytosolic DNA induces interferons through the production of cyclic guanosine monophosphate-adenosine monophosphate (cyclic GMP-AMP, or cGAMP), which binds to and activates the adaptor protein STING. Through biochemical fractionation and quantitative mass spectrometry, we identified a cGAMP synthase (cGAS), which belongs to the nucleotidyltransferase family. Overexpression of cGAS activated the transcription factor IRF3 and induced interferon-β in a STING-dependent manner. Knockdown of cGAS inhibited IRF3 activation and interferon-β induction by DNA transfection or DNA virus infection. cGAS bound to DNA in the cytoplasm and catalyzed cGAMP synthesis. These results indicate that cGAS is a cytosolic DNA sensor that induces interferons by producing the second messenger cGAMP.
Figures
Figure 1. Identification of a cGAMP synthase (cGAS)
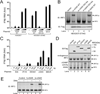
(A) Multiple sequence and structure alignment of putative nucleotidyltransferase (NTase) domain of mouse cGAS, human cGAS and human OAS1 using the PROMALS3D program. Conserved active site residues of NTase superfamily are highlighted in black, identical amino acids in red and conserved amino acids in yellow. Predicted secondary structure is indicated above the alignment as alpha helices (H) and beta strands (E). (B and C) Quantitative RT-PCR analyses of cGAS RNA levels in different murine (B) and human (C) cell lines. MEF (imt): immortalized MEF; Raw: Raw264.7; SDM: spleen-derived macrophage; BMDM: bone marrow-derived macrophage. The error bars in this and all other q-RT-PCR assays represent standard errors of the mean (n=3). (D) Immunoblotting of endogenous human proteins in HEK293T and THP1 cells with the indicated antibodies.
Figure 2. cGAS activates IRF3 and induces IFNβ
(A) Expression plasmids (100 and 500 ng) encoding Flag-tagged mouse cGAS (m-cGAS), its active site mutants G198A/S199A (designated as GS>AA), and MAVS were transfected into HEK293T cells or the same cell line stably expressing STING (HEK293T-STING). 24 hr after transfection, IFNβ RNA was measured by q-RT-PCR. (B) Similar to (A), except that cell lysates were analyzed for IRF3 dimerization by native gel electrophoresis (top). The expression levels of the transfected genes were monitored by immunoblotting with a Flag antibody (bottom). h-cGAS: human cGAS; ED>AA: E211A/D213A in mouse cGAS. (C) Expression vectors for the indicated proteins were transfected into HEK293T-STING cells, followed by measurement of IFNβ by q-RT-PCR. (D) Cell lysates shown in (C) were immunoblotted with antibodies against Flag and IRF3 after SDS-PAGE and native PAGE, respectively (top two panels). Aliquots of the cell extracts were assayed for the presence of cGAMP activity, which was measured by detecting IRF3 dimerization after delivery into permeabilized Raw264.7 cells (bottom). (E) Human and mouse cGAS were expressed in HEK293T cells and affinity purified using a Flag antibody. The proteins were incubated with ATP and GTP in the presence or absence of HT-DNA, and the synthesis of cGAMP was assessed by its ability to induce IRF3 dimerization in Raw264.7 cells.
Figure 3. cGAS is essential for IRF3 activation and IFNβ induction by DNA transfection and DNA virus infection
(A) L929 cell lines stably expressing shRNA targeting GFP (control) or two different regions of m-cGAS were transfected with HT-DNA for indicated times, followed by measurement of IFNβ RNA by q-RT-PCR. See fig. S4B for RNAi efficiency. (B) L929 cells stably expressing shRNA against GFP, cGAS or STING were transfected with pcDNA3 (vector) or the same vector driving the expression of indicated proteins. 24 hr after transfection, IFNβ RNA was measured by q-RT-PCR. see fig. S4C for RNAi efficiency. (C) cGAMP (100 nM) was delivered to digitoninpermeabilized L929/shRNA cells as indicated. IFNβ RNA was measured by q-RT-PCR at indicated times following cGAMP delivery. (D and E) L929-shRNA cells as indicated were infected with HSV1 (ΔICP34.5) or Sendai virus (SeV) for indicated times followed by measurement of IRF3 dimerization. (F) L929/shRNA cells were transfected with HT-DNA or infected with HSV1 for 6 hr, followed by measurement of IRF3 dimerization (top). Extracts from these cells were used to prepare heat-resistant supernatants, which were delivered to permeabilized Raw264.7 cells to stimulate IRF3 dimerization (bottom). (G) The heat-resistant supernatants in (F) were fractionated by HPLC using a C18 column and the abundance of cGAMP was quantitated by mass spectrometry using SRM.
Figure 4. DNA-dependent synthesis of cGAMP by purified cGAS
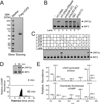
(A) Silver staining of Flag-h-cGAS expressed and purified from HEK293T cells. (B) Purified Flag-h-cGAS as shown in (A) was incubated with ATP and GTP, in the presence of different forms of nucleic acids as indicated. Generation of cGAMP was assessed by its ability to induce IRF3 dimerization in Raw264.7 cells. (C) Similar to (B), except that reactions contained HT-DNA and different combinations of NTP as indicated. (D) Similar to (B), except that WT and mutant cGAS proteins were expressed and purified from E. coli and assayed for their activities at indicated concentrations. (E) Purified m-cGAS from E coli was incubated with ATP, GTP and DNA for 0 or 60 min, and the production of cGAMP was analyzed by IRF3 dimerization assay (top) and mass spectrometry using SRM (bottom).
Figure 5. cGAS is a DNA binding protein
(A) Indicated GST fusion proteins were expressed and purified from E. coli and then incubated with Streptavidin beads in the presence of ISD or biotin-ISD. Bound proteins were eluted with SDS sample buffer and detected by immunoblotting with a GST antibody. (B) Flag-h-cGAS was expressed and purified from HEK293T cells and then incubated with streptavidin beads as described in (A) except that a Flag antibody was used in immunoblotting and a biotin-RNA was also tested for binding to cGAS. (C) Flag-tagged full-length or truncated human cGAS proteins were expressed in HEK293T cells and affinity purified. Their ability to bind biotin-ISD was assayed as described in (B). Right panel: Expression plasmids encoding full-length and deletion mutants of h-cGAS were transfected into HEK293T-STING cells followed by measurement of IFNβ RNA by q-RT-PCR.
Figure 6. cGAS binds to DNA in the cytoplasm
(A) Nuclear and cytoplasmic fractions were prepared from THP-1 cells and analyzed by immunoblotting with the indicated antibodies. (B) THP-1 cells were homogenized in hypotonic buffer and subjected to differential centrifugation. Pellets at different speeds of centrifugation (e.g, P100: pellets after 100,000 × g) and S100 were immunoblotted with the indicated antibodies. (C) L929 cells stably expressing Flag-cGAS (green) were transfected with Cy3-ISD (red). At different time points after tranfection, cells were fixed, stained with the Flag antibody or DAPI and imaged by confocal fluorescence microscopy. Inset: magnification of the area outlined in the merged images. These images are representative of at least 10 cells at each time point (representing > 50% of the cells under examination).
Comment in
- Immunology. Sensing the dark side of DNA.
O'Neill LA. O'Neill LA. Science. 2013 Feb 15;339(6121):763-4. doi: 10.1126/science.1234724. Science. 2013. PMID: 23413341 No abstract available.
Similar articles
- Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA.
Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Wu J, et al. Science. 2013 Feb 15;339(6121):826-30. doi: 10.1126/science.1229963. Epub 2012 Dec 20. Science. 2013. PMID: 23258412 Free PMC article. - The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop.
Zhang X, Wu J, Du F, Xu H, Sun L, Chen Z, Brautigam CA, Zhang X, Chen ZJ. Zhang X, et al. Cell Rep. 2014 Feb 13;6(3):421-30. doi: 10.1016/j.celrep.2014.01.003. Epub 2014 Jan 23. Cell Rep. 2014. PMID: 24462292 Free PMC article. - Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses.
Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Gao D, et al. Science. 2013 Aug 23;341(6148):903-6. doi: 10.1126/science.1240933. Epub 2013 Aug 8. Science. 2013. PMID: 23929945 Free PMC article. - Cyclic GMP-AMP as an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA.
Kato K, Omura H, Ishitani R, Nureki O. Kato K, et al. Annu Rev Biochem. 2017 Jun 20;86:541-566. doi: 10.1146/annurev-biochem-061516-044813. Epub 2017 Apr 7. Annu Rev Biochem. 2017. PMID: 28399655 Review. - cGAS and cancer therapy: a double-edged sword.
Du JM, Qian MJ, Yuan T, Chen RH, He QJ, Yang B, Ling Q, Zhu H. Du JM, et al. Acta Pharmacol Sin. 2022 Sep;43(9):2202-2211. doi: 10.1038/s41401-021-00839-6. Epub 2022 Jan 18. Acta Pharmacol Sin. 2022. PMID: 35042992 Free PMC article. Review.
Cited by
- Mechanism and Application Prospects of NLRC3 Regulating cGAS-STING Pathway in Lung Cancer Immunotherapy.
Wang Q, Ren Z, Zhao J, Zheng T, Tong L, Liu J, Dai Z, Tang S. Wang Q, et al. Int J Med Sci. 2024 Oct 7;21(13):2613-2622. doi: 10.7150/ijms.102328. eCollection 2024. Int J Med Sci. 2024. PMID: 39439455 Free PMC article. Review. - Nucleic acid recognition orchestrates the anti-viral response to retroviruses.
Stavrou S, Blouch K, Kotla S, Bass A, Ross SR. Stavrou S, et al. Cell Host Microbe. 2015 Apr 8;17(4):478-88. doi: 10.1016/j.chom.2015.02.021. Epub 2015 Mar 26. Cell Host Microbe. 2015. PMID: 25816774 Free PMC article. - Circulating Cell-Free DNA Level in Prediction of COVID-19 Severity and Mortality: Correlation of with Haematology and Serum Biochemical Parameters.
Mishra S, Dubey DB, Agarwal K, Dubey DB, Verma S, Shabbir N, Kushwaha R, Reddy DH, Singh US, Ali W. Mishra S, et al. Indian J Clin Biochem. 2023 Apr;38(2):172-181. doi: 10.1007/s12291-022-01082-4. Epub 2022 Aug 21. Indian J Clin Biochem. 2023. PMID: 36032561 Free PMC article. - 2024 Lasker Award Recipient Zhijian Chen elucidates how DNA stimulates immunity.
Heimberger AB. Heimberger AB. J Clin Invest. 2024 Sep 19;134(19):e186104. doi: 10.1172/JCI186104. J Clin Invest. 2024. PMID: 39295301 Free PMC article. No abstract available. - QnAs with Zhijian "James" Chen: Winner of the 2024 Albert Lasker Basic Medical Research Award.
Hardcastle M. Hardcastle M. Proc Natl Acad Sci U S A. 2024 Sep 24;121(39):e2415810121. doi: 10.1073/pnas.2415810121. Epub 2024 Sep 19. Proc Natl Acad Sci U S A. 2024. PMID: 39297684 Free PMC article. No abstract available.
References
- O'Neill LA. DNA makes RNA makes innate immunity. Cell. 2009 Aug 7;138:428. - PubMed
- Barber GN. Cytoplasmic DNA innate immune pathways. Immunological reviews. 2011 Sep;243:99. - PubMed
- Keating SE, Baran M, Bowie AG. Cytosolic DNA sensors regulating type I interferon induction. Trends in immunology. 2011 Dec;32:574. - PubMed
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008 Dec;26:1367. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials