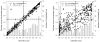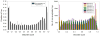Protein intrinsic disorder in the acetylome of intracellular and extracellular Toxoplasma gondii - PubMed (original) (raw)
Protein intrinsic disorder in the acetylome of intracellular and extracellular Toxoplasma gondii
Bin Xue et al. Mol Biosyst. 2013.
Abstract
Toxoplasma gondii is an obligate intracellular parasite of the phylum Apicomplexa, which includes a number of species of medical and veterinary importance. Inhibitors of lysine deacetylases (KDACs) exhibit potent antiparasitic activity, suggesting that interference with lysine acetylation pathways holds promise for future drug targeting. Using high resolution LC-MS/MS to identify parasite peptides enriched by immunopurification with acetyl-lysine antibody, we recently produced an acetylome of the proliferative intracellular stage of Toxoplasma. In this study, we used similar approaches to greatly expand the Toxoplasma acetylome by identifying acetylated proteins in non-replicating extracellular tachyzoites. The functional breakdown of acetylated proteins in extracellular parasites is similar to intracellular parasites, with an enrichment of proteins involved in metabolism, translation, and chromatin biology. Altogether, we have now detected over 700 acetylation sites on a wide variety of parasite proteins of diverse function in multiple subcellular compartments. We found 96 proteins uniquely acetylated in intracellular parasites, 216 uniquely acetylated in extracellular parasites, and 177 proteins acetylated in both states. Our findings suggest that dramatic changes occur at the proteomic level as tachyzoites transition from the intracellular to the extracellular environment, similar to reports documenting significant changes in gene expression during this transition. The expanded dataset also allowed a thorough analysis of the degree of protein intrinsic disorder surrounding lysine residues targeted for this post-translational modification. These analyses indicate that acetylated lysines in proteins from extracellular and intracellular tachyzoites are largely located within similar local environments, and that lysine acetylation preferentially occurs in intrinsically disordered or flexible regions.
Figures
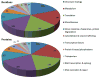
Figure 1. Overview of the extracellular tachyzoite acetylome
Lysine acetylation occurs on proteins associated with virtually every cellular process, but the majority of acetylation occurs on proteins involved in metabolism, translation, and chromatin biology. The pie chart on the right shows the functional categorization of the 571 acetylated residues found across the 386 acetylated proteins, which are displayed in the left pie chart. Acetylation of the 172 lysines across 125 hypothetical proteins was omitted in these pie charts.
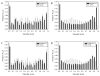
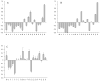
Figure 2. Comparison of the disorder status of acetylated and non-acetylated lysines in the extracellular and intracellular tachyzoites
Here, the distributions of lysines over the various ranges of disorder score are shown. Comparison of the outputs of the PONDR® VLXT (black bars) and IUPred (gray bars) for the acetylated (A) and non-acetylated (B) lysines in the extracellular tachyzoites, and for the acetylated (C) and non-acetylated (D) lysines in the intracellular tachyzoites. Here, the distributions of lysines over the various ranges of disorder score are shown for different datasets. Error bars corresponds to the errors evaluated by bootstrapping.
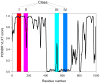
Figure 3. Classification of lysines based on their local environment within the sequences of parent proteins
Class I, a disordered lysine is located inside a disordered region; Class II, an ordered lysine is located inside a disordered region; Class III, a disordered lysine is located inside an ordered region; and Class IV, an ordered lysine is located inside an ordered region. PONDR® VLXT curve of a model protein is shown and positions of mentioned classes are shown as red, pink, cyan, and blue shaded areas. This plot contains a model disorder profile to illustrate principle of analysis.
Figure 4. Disorder scores of acetylated lysines and analysis of their local environment
The disorder score of each acetylated lysine in the 274 proteins from intracellular tachyzoites was predicted using PONDR® VLXT. The influence of neighboring residues was measured by averaging disorder scores over peptides centered at the target lysine but extended on both sides by a specific number of residues. The length of the extension is 5 (A) or 30 residues (B) and the corresponding scores are labeled as vlxt-5 and vlxt-30; vlxt-0 represents the original prediction score without an extension. Each “x” in the figure is an acetylated lysine. The x-axis displays the original disorder scores for the acetylated lysines and the y-axis displays the average disorder score over the extended regions. The gray bars represent the distribution of the original PONDR® VLXT scores for the acetylated lysines. Similar plots generated for vlxt-10, vlxt-15, vlxt-20, and vlxt-25 are shown in Supplementary Figure S2.
Figure 5. Analysis of the intrinsic disorder status of acetylated and non-acetylated lysines in intracellular tachyzoites
A. Distribution of acetylated (gray bars) and non-acetylated lysines (black bars) over the various ranges of disorder score. Each bar represents the fraction of acetylated or non-acetylated lysines for a given disorder score evaluated by PONDR® VLXT. Error bars corresponds to the errors evaluated by bootstrapping. B. Analysis of the local environment of non-acetylated lysines within the amino acid sequences of 274 tachyzoite proteins represented as changes in the averaged PONDR® VLXT score distribution for the non-acetylated lysines. Extension-0 is from the original PONDR® VLXT score. Extension-N corresponds to the averaged disorder score calculated for a lysine-containing region extended by N amino acids at each termini.
Figure 6. Analysis of sequences flanking lysines targeted for acetylation
Logo representation of the amino acids flanking acetylated (A) and non-acetylated (B) lysines in 274 proteins from intracellular tachyzoites. Residues are colored according to their disorder propensities – disorder-promoting (red), order-promoting (blue), neutral (green).
Figure 7. Composition profiling of 15 residues-long peptides containing acetylated lysines (A) or non-acetylated lysines (B) in comparison with ordered proteins
Both types of peptides are clearly disordered since they are depleted in major order-promoting residues and are enriched in several disorder-promoting residues. However, the amino acid compositions of these two types of regions are clearly different. In these plots, y-axis represents (Cs1 − Cs2)/Cs2 values calculated for each residue shown in x-axis, where Cs1 is a content of a given residue in a query set of peptides (15 residues-long peptides containing acetylated lysines or non-acetylated lysines), whereas Cs2 is the corresponding value for the sample set of proteins (a set of ordered proteins from PDB). Negative bars correspond to the residues which are under-represented in set S1, whereas positive bars correspond to the over-represented residues. Error bars represent the non-parametric estimations of the confidence intervals for the reported amino acid compositions. These confidence intervals are standard deviations of the pseudo-replicate compositions calculated by bootstrapping (1,000 iterations), where in each iteration step, random samples of the two starting samples are created, relative entropy between the random samples is computed and compared to the observed relative entropy. (C) Compositional profiling of peptides with non-acetylated lysines in comparison with the peptides containing acetylated lysines. Acetylated peptides clearly have more of C, F, I, Y and G (statistically significant enrichment is for C, F, Y and G) and less of W, M, R, P and E (statistically significant depletion is for R, P and E). Here, y-axis represents (Cs1 − Cs2)/Cs2 values calculated for each residue shown in x-axis, where Cs1 is a content of a given residue in a set of peptides containing non-acetylated lysines, whereas Cs2 is the corresponding value for the set of set of peptides containing acetylated lysines proteins.
Figure 8. Comparison of the local environments of acetylated sites in extracellular (A) and intracellular tachyzoites (B)
Similar to the data shown in Figure 4, the influence of neighboring residues was measured by averaging disorder scores over peptides centered at the target lysine but extended on both sides by a specific number of residues. The length of the extension is 15 and the corresponding score is labeled as vlxt-15; vlxt-0 represents the original prediction score without an extension. Each symbol in the figure is an acetylated lysine. The x-axis displays the original disorder scores for the acetylated lysines and the y-axis displays the average disorder score over the extended regions.
Figure 9. Disordered nature of the acetylation sites
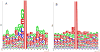
A. Crystal structure (PDB ID: 2P0W) of a complex between human histone acetyltransferase 1 (HAT1, shown as a cyan cloud) and a 15-amino acid peptide derived from the human histone H4 (depicted in red). B. Crystal structure (PDB ID: 1PU9) of a complex between the Tetrahymena thermophila histone acetyltransferase GCN5 (cyan cloud) and a 19-amino acid peptide derived from the histone H3 (red). C. Crystal structure (PDB ID: 3Q33) of a complex between the histone acetyltransferase RTT109 from Saccharomyces cerevisiae (blue cloud), a 14-amino acid peptide from histone H3 (red, note that only 4 residues were resolved, whereas 10 residues were missing in the electron density map) and a histone chaperone from Saccharomyces cerevisiae (vacuolar protein sorting-associated protein 75, shown as a cyan cloud). D. NMR solution structure (PDB ID: 2RNW) of a complex between a bromodomain of the human acetyltransferase PCAF (cyan cloud shows one member of the conformational ensemble) and 14-amino acid peptide from histone H3 (10 members of the structural ensemble are shown by strands of different color).
Similar articles
- Lysine acetylation is widespread on proteins of diverse function and localization in the protozoan parasite Toxoplasma gondii.
Jeffers V, Sullivan WJ Jr. Jeffers V, et al. Eukaryot Cell. 2012 Jun;11(6):735-42. doi: 10.1128/EC.00088-12. Epub 2012 Apr 27. Eukaryot Cell. 2012. PMID: 22544907 Free PMC article. - Systematic identification of the lysine succinylation in the protozoan parasite Toxoplasma gondii.
Li X, Hu X, Wan Y, Xie G, Li X, Chen D, Cheng Z, Yi X, Liang S, Tan F. Li X, et al. J Proteome Res. 2014 Dec 5;13(12):6087-95. doi: 10.1021/pr500992r. Epub 2014 Nov 17. J Proteome Res. 2014. PMID: 25377623 - Proteome-wide lysine acetylation in cortical astrocytes and alterations that occur during infection with brain parasite Toxoplasma gondii.
Bouchut A, Chawla AR, Jeffers V, Hudmon A, Sullivan WJ Jr. Bouchut A, et al. PLoS One. 2015 Mar 18;10(3):e0117966. doi: 10.1371/journal.pone.0117966. eCollection 2015. PLoS One. 2015. PMID: 25786129 Free PMC article. - Stage differentiation of the protozoan parasite Toxoplasma gondii.
Bohne W, Holpert M, Gross U. Bohne W, et al. Immunobiology. 1999 Dec;201(2):248-54. doi: 10.1016/S0171-2985(99)80065-5. Immunobiology. 1999. PMID: 10631574 Review. - Post-translational modifications as key regulators of apicomplexan biology: insights from proteome-wide studies.
Yakubu RR, Weiss LM, Silmon de Monerri NC. Yakubu RR, et al. Mol Microbiol. 2018 Jan;107(1):1-23. doi: 10.1111/mmi.13867. Epub 2017 Nov 28. Mol Microbiol. 2018. PMID: 29052917 Free PMC article. Review.
Cited by
- Elongator protein 3 (Elp3) lysine acetyltransferase is a tail-anchored mitochondrial protein in Toxoplasma gondii.
Stilger KL, Sullivan WJ Jr. Stilger KL, et al. J Biol Chem. 2013 Aug 30;288(35):25318-25329. doi: 10.1074/jbc.M113.491373. Epub 2013 Jul 22. J Biol Chem. 2013. PMID: 23878194 Free PMC article. - Proteome and Acetyl-Proteome Profiling of Camellia sinensis cv. 'Anjin Baicha' during Periodic Albinism Reveals Alterations in Photosynthetic and Secondary Metabolite Biosynthetic Pathways.
Xu YX, Chen W, Ma CL, Shen SY, Zhou YY, Zhou LQ, Chen L. Xu YX, et al. Front Plant Sci. 2017 Dec 11;8:2104. doi: 10.3389/fpls.2017.02104. eCollection 2017. Front Plant Sci. 2017. PMID: 29312376 Free PMC article. - A latent ability to persist: differentiation in Toxoplasma gondii.
Jeffers V, Tampaki Z, Kim K, Sullivan WJ Jr. Jeffers V, et al. Cell Mol Life Sci. 2018 Jul;75(13):2355-2373. doi: 10.1007/s00018-018-2808-x. Epub 2018 Mar 30. Cell Mol Life Sci. 2018. PMID: 29602951 Free PMC article. Review. - Dynamic Protein Interaction Networks and New Structural Paradigms in Signaling.
Csizmok V, Follis AV, Kriwacki RW, Forman-Kay JD. Csizmok V, et al. Chem Rev. 2016 Jun 8;116(11):6424-62. doi: 10.1021/acs.chemrev.5b00548. Epub 2016 Feb 29. Chem Rev. 2016. PMID: 26922996 Free PMC article. Review. - Dynamic Protein Acetylation in Plant-Pathogen Interactions.
Song G, Walley JW. Song G, et al. Front Plant Sci. 2016 Mar 30;7:421. doi: 10.3389/fpls.2016.00421. eCollection 2016. Front Plant Sci. 2016. PMID: 27066055 Free PMC article. Review.
References
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Science. 2009;325:834–840. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases