MiR-181a regulates inflammation responses in monocytes and macrophages - PubMed (original) (raw)
MiR-181a regulates inflammation responses in monocytes and macrophages
Weidong Xie et al. PLoS One. 2013.
Abstract
miR-181a has been presumed to target the 3'-untranslated regions (3'-UTR) of IL1a based on software predictions. miR-181a and IL1a have opposite expression levels in monocytes and macrophages in the inflammatory state. This led us to suspect that mir-181a has an important function in regulating inflammatory response by targeting IL1a. Fluorescence reporter assays showed that miR-181a effectively binds to the 3'-UTR of IL1a. The anti-inflammatory functions of miR-181a were investigated in lipopolysaccharides (LPS)-induced Raw264.7 and phorbol 12-myristate 13-acetate (PMA)/LPS-induced THP-1 cells. We found that miR-181a mimics significantly lowered IL1a expression levels in these cells and, interestingly, miR-181a inhibitors reversed this decrease. In addition, miR-181a mimics significantly inhibited increase in the levels of inflammatory factors (IL1b, IL6, and TNFa) in these cells. Furthermore, miR-181a mimics and inhibitors decreased and increased, respectively, production of reactive oxygen species in PMA/LPS-induced THP-1 cells. These results indicate that miR-181a regulates inflammatory responses by directly targeting the 3'-UTR of IL1a and down-regulating IL1a levels. Interestingly, we found that miR-181a inhibited production of inflammatory factors even in IL1a-induced THP-1 cells, suggesting that the anti-inflammatory effects of miR-181a possibly involves other targets in addition to IL1a. Thus, we provide the first evidence for anti-inflammatory effects of miR-181a mediated at least in part by down-regulating IL1a.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Presumed binding sites for miR-181a in the 3′-UTR of mouse and human IL1a.
The binding sites are highly conserved.
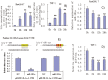
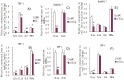
Figure 2. (A and B) miR-181a expression in LPS-induced Raw264.7 cells and PMA/LPS-induced THP-1 cells; (C and D) intracellular IL1a levels in LPS-induced Raw264.7 cells and PMA/LPS-induced THP-1 cells; (E) effects of miR-181a mimics on chemiluminescence values in Cos-7 cells (transfected with pRLTK containing L1a-3′-UTR or its mutated variant).
Data are expressed as mean ± SD (n = 3), **P<0.01 vs. values at 0 h (A–D), **P<0.01 vs. negative control (NC, E).
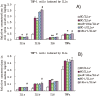
Figure 3. Effects of miR-181a mimics and inhibitors on IL1a and IL1b mRNA levels in LPS-induced Raw264.7 (A) and PMA/LPS-induced THP-1 cells (B); effects of miR-181a mimics and inhibitors on ILa, IL1b, IL6, and TNFa in cell lysates and media of LPS-induced Raw264.7 (C and E) and PMA/LPS-induced THP-1 cells (D and F) assayed by ELISA.
Data are expressed as mean ± SD (n = 3), **P<0.01 vs. negative control (NC), # P<0.05, ## P<0.01 vs. miRNA inhibitor negative control (NC-i). ‘NC-i’ is a single stranded nucleic acid used as negative control for miR-181a inhibitors (181a–i). ‘LPS + or −’, cells were treated with or without LPS in the case of Raw264.7 cells or PMA/LPS in the case of THP-1 cells.
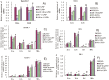
Figure 4. Effects of miR-181a mimics and inhibitors on ILa levels in cell lysis solution of LPS-induced Raw264.7 (A) and PMA/LPS-induced THP-1 cells (B) assayed by Western blot.
Data are expressed as mean ± SD (n = 3–5), **P<0.01 vs. negative control (NC), ## P<0.01 vs miRNA inhibitor negative control (NC-i). ‘NC-i’ is a single-stranded nucleic acid used as negative control for miR-181a inhibitors (181a–i). Effects of miR-181a precursors (pre-miR-181a) and its inhibitors on ILa, IL1b, IL6, and TNFa in cell lysates and culture media of PMA/LPS-induced THP-1 cells (C and D) assayed by ELISA. Data are expressed as mean ± SD (n = 3), *P<0.05, **P<0.01 vs. negative control (pre-NC), # P<0.05 vs. miRNA inhibitor negative control (pre-NC-i). ‘pre-NC-i’ is a single-stranded nucleic acid used as negative control for miR-181a inhibitors (pre-miR-181a–i). ‘LPS + or −’, cells were treated with or without LPS in the case of Raw264.7 cells or PMA/LPS in the case of THP-1 cells.
Figure 5. Effects of stable expression of miR-181a on production of inflammatory factors in LPS-induced Raw264.7 cells.
(A) pCMV recombinant Raw264.7 cells (CMV, negative controls); (B) pCMV-pre-miR-181a recombinant Raw264.7 cells (SC); (C) endogenous miR-181a expression in SC (containing miR-181a expression vector) and CMV (containing negative control vector) cells; (D) Expression of IL1a in pCMV and pCMV-pre-miR-181a recombinant Raw264.7 cells assayed by Western blotting; (E and F) Levels of IL1a, IL1b, IL6, and TNFa in cell lysis solution and culture media of recombinant Raw264.7 cells transfected with pCMV and pCMV-pre-miR-181a. Data are expressed as mean ± SD (n = 3), *P<0.05, **P<0.01 vs. pCMV recombinant controls induced by LPS (CMV LPS+), ## P<0.01 vs. miRNA inhibitor negative control (NC-i). LPS+ and LPS− indicate that cells were treated with and without LPS, respectively.
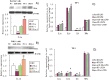
Figure 6. (A and B) Effects of transient transfection with miR-181a expression vector on the levels of ILa, IL1b, IL6, and TNFa in PMA/LPS-induced THP-1 cell lysis solution and culture media.
Data are expressed as mean ± SD (n = 3), *P<0.05, **P<0.01 vs. CMV controls. ‘SC’, pCMV-pre-miR-181a recombinant THP-1 cells; ‘CMV’, pCMV recombinant THP-1 cells (negative controls). (C and D) Effects of si-IL1a on the mRNA levels of IL1a and IL1b in LPS-induced Raw264.7 and PMA/LPS-induced THP-1 cells. (E and F) Effects of si-IL1a on the levels of IL1a, IL1b, IL6, and TNFa in LPS-induced Raw264.7 and PMA/LPS-induced THP-1 cells. Data are expressed as mean ± SD (n = 3), *P<0.05, **P<0.01 vs. negative control (NC). ‘siNC’ is a single-stranded nucleic acid used as negative control for siIL1a.
Figure 7. Effects of miR-181a mimics and inhibitors on the levels of IL1a, IL1b, IL6, and TNFa in IL1a-induced THP-1 cell lysis solution (A) and culture medium (B).
Data are expressed as mean ± SD (n = 3), *P<0.05, **P<0.01 vs. negative control (NC), # P<0.05 vs. miRNA inhibitor negative control (NC-i). ‘NC-i’ is a single-stranded nucleic acid used as a negative control for miR-181a inhibitors (181a–i). IL1a + and IL1a- indicate that the cells were treated with (at a final concentration of 0.1 ng/ml) and without exogenous IL1a, respectively.
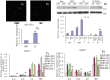
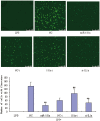
Figure 8. Effects of miR-181a mimics and inhibitors on the productions of reactive oxygen species (ROS) in PMA/LPS-induced TPH-1 cells.
Data are expressed as mean ± SD (n = 3), **P<0.01 vs. negative control (NC), ## P<0.01 vs. miRNA inhibitor negative control (NC-i). ‘NC-i’ is a single-stranded nucleic acid used as negative control for miR-181a inhibitors (181a–i). LPS+ and LPS− indicate cells treated with and without PMA/LPS, respectively.
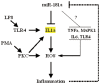
Figure 9. Potential effects and mechanisms of the regulation of inflammation by miR-181a.
Similar articles
- Regulatory roles of miR-155 and let-7b on the expression of inflammation-related genes in THP-1 cells: effects of fatty acids.
Marques-Rocha JL, Garcia-Lacarte M, Samblas M, Bressan J, Martínez JA, Milagro FI. Marques-Rocha JL, et al. J Physiol Biochem. 2018 Nov;74(4):579-589. doi: 10.1007/s13105-018-0629-x. Epub 2018 May 22. J Physiol Biochem. 2018. PMID: 29790117 - miR-181a and inflammation: miRNA homeostasis response to inflammatory stimuli in vivo.
Xie W, Li Z, Li M, Xu N, Zhang Y. Xie W, et al. Biochem Biophys Res Commun. 2013 Jan 11;430(2):647-52. doi: 10.1016/j.bbrc.2012.11.097. Epub 2012 Dec 5. Biochem Biophys Res Commun. 2013. PMID: 23220232 - MiR-34a inhibits lipopolysaccharide-induced inflammatory response through targeting Notch1 in murine macrophages.
Jiang P, Liu R, Zheng Y, Liu X, Chang L, Xiong S, Chu Y. Jiang P, et al. Exp Cell Res. 2012 Jun 10;318(10):1175-84. doi: 10.1016/j.yexcr.2012.03.018. Epub 2012 Mar 27. Exp Cell Res. 2012. PMID: 22483937 - miR-130b-3p regulates M1 macrophage polarization via targeting IRF1.
Guo Q, Zhu X, Wei R, Zhao L, Zhang Z, Yin X, Zhang Y, Chu C, Wang B, Li X. Guo Q, et al. J Cell Physiol. 2021 Mar;236(3):2008-2022. doi: 10.1002/jcp.29987. Epub 2020 Aug 27. J Cell Physiol. 2021. PMID: 32853398 - MiR-145 improves macrophage-mediated inflammation through targeting Arf6.
Li R, Shen Q, Wu N, He M, Liu N, Huang J, Lu B, Yao Q, Yang Y, Hu R. Li R, et al. Endocrine. 2018 Apr;60(1):73-82. doi: 10.1007/s12020-018-1521-8. Epub 2018 Jan 31. Endocrine. 2018. PMID: 29388044
Cited by
- The RNA binding protein IGF2BP2/IMP2 alters the cargo of cancer cell-derived extracellular vesicles supporting tumor-associated macrophages.
Mashayekhi V, Schomisch A, Rasheed S, Aparicio-Puerta E, Risch T, Yildiz D, Koch M, Both S, Ludwig N, Legroux TM, Keller A, Müller R, Fuhrmann G, Hoppstädter J, Kiemer AK. Mashayekhi V, et al. Cell Commun Signal. 2024 Jun 27;22(1):344. doi: 10.1186/s12964-024-01701-y. Cell Commun Signal. 2024. PMID: 38937789 Free PMC article. - Molecular Pathogenesis of Ischemic and Hemorrhagic Strokes: Background and Therapeutic Approaches.
Maida CD, Norrito RL, Rizzica S, Mazzola M, Scarantino ER, Tuttolomondo A. Maida CD, et al. Int J Mol Sci. 2024 Jun 7;25(12):6297. doi: 10.3390/ijms25126297. Int J Mol Sci. 2024. PMID: 38928006 Free PMC article. Review. - Exosome Content-Mediated Signaling Pathways in Multiple Sclerosis.
Mohammadinasr M, Montazersaheb S, Ayromlou H, Hosseini V, Molavi O, Hejazi MS. Mohammadinasr M, et al. Mol Neurobiol. 2024 Aug;61(8):5404-5417. doi: 10.1007/s12035-023-03862-2. Epub 2024 Jan 8. Mol Neurobiol. 2024. PMID: 38191693 Review. - Circulating cell-free nucleic acids of plasma in human aging, healthy aging and longevity: current state of knowledge.
Tessier NP, Hardy LM, Deleuze JF, How-Kit A. Tessier NP, et al. Front Genet. 2023 Nov 28;14:1321280. doi: 10.3389/fgene.2023.1321280. eCollection 2023. Front Genet. 2023. PMID: 38090154 Free PMC article. Review. - MicroRNAs: Small but Key Players in Viral Infections and Immune Responses to Viral Pathogens.
Bauer AN, Majumdar N, Williams F, Rajput S, Pokhrel LR, Cook PP, Akula SM. Bauer AN, et al. Biology (Basel). 2023 Oct 14;12(10):1334. doi: 10.3390/biology12101334. Biology (Basel). 2023. PMID: 37887044 Free PMC article. Review.
References
- Xie W, Du L (2011) Diabetes is an inflammatory disease: evidences from traditional Chinese Medicines. Diabetes Obes Metab 13: 289–301. - PubMed
- Taube A, Schlich R, Sell H, Eckardt K, Eckel J (2012) Inflammation and metabolic dysfunction: links to cardiovascular diseases. Am J Physiol Heart Circ Physiol 302: H2148–2165. - PubMed
- Vendramini-Costa DB, Carvalho JE (2012) Molecular Link Mechanisms between Inflammation and Cancer. Curr Pharm Des 18: 3831–3852. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This study was supported by the National Natural Science Foundation of China (81072680), the Guangdong Natural Science Foundation (10151805702000002), the Specialized Research Fund for the Doctoral Program of Higher Education of China (20100002120017), and the Shenzhen Science and Technology R&D Foundation (ZYC201105170341A). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases