mir-17-92: a polycistronic oncomir with pleiotropic functions - PubMed (original) (raw)
Review
mir-17-92: a polycistronic oncomir with pleiotropic functions
Virginie Olive et al. Immunol Rev. 2013 May.
Abstract
Neoplastic transformation is caused by accumulation of genetic lesions that ultimately convert normal cells into tumor cells with uncontrolled proliferation and survival, unlimited replicative potential, and invasive growth. Emerging evidence has highlighted the functional importance of non-coding RNAs, particularly microRNAs (miRNAs), in the initiation and progression of tumor development. The mir-17-92 miRNA is among the best characterized miRNA oncogenes, whose genomic amplification or aberrant elevation are frequently observed in a variety of tumor types. Unlike protein-coding oncogenes, where one transcript produces one protein, mir-17-92 encodes a polycistronic miRNA transcript that yields six individual miRNA components. This unique gene structure, shared by many important miRNA oncogenes and tumor suppressors, underlies the unique functionality of mir-17-92 in a cell type and context-dependent manner. Recent functional dissection of mir-17-92 indicates that individual mir-17-92 components perform distinct biological functions, which collectively regulate multiple related cellular processes during development and disease. The structural complexity of mir-17-92 as a polycistronic miRNA oncogene, along with the complex mode of interactions among its components, constitutes the molecular basis for its unique functional complexity during normal and tumor development.
© 2013 John Wiley & Sons A/S. Published by Blackwell Publishing Ltd.
Figures
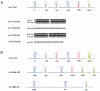
Fig. 1
mir-17-92 encodes a polycistronic miRNA oncogene. A. The gene structure of the mir-17-92 polycistron. mir-17-92 encodes a miRNA precursor that yields 6 mature miRNAs. Based on seed sequence homology, these six miRNAs belong to four miRNA families. Homologous miRNA components are indicated by the same color. B. mir-17-92 has two closely related homologues in mammals. The mir-106a-393 and mir-106b-25 clusters contain homologous miRNAs to a subset of mir-17-92 components with a conserved genome arrangement.
Fig. 2
A diagram of possible regulatory mechanisms that collectively modulate the biological functions of the mir-17-92 polycistronic oncomir.
Similar articles
- mir-17-92, a cluster of miRNAs in the midst of the cancer network.
Olive V, Jiang I, He L. Olive V, et al. Int J Biochem Cell Biol. 2010 Aug;42(8):1348-54. doi: 10.1016/j.biocel.2010.03.004. Epub 2010 Mar 19. Int J Biochem Cell Biol. 2010. PMID: 20227518 Free PMC article. Review. - MicroRNAs: small but potent oncogenes or tumor suppressors.
Lee YS, Dutta A. Lee YS, et al. Curr Opin Investig Drugs. 2006 Jun;7(6):560-4. Curr Opin Investig Drugs. 2006. PMID: 16784027 Review. - An expanding universe of the non-coding genome in cancer biology.
Xue B, He L. Xue B, et al. Carcinogenesis. 2014 Jun;35(6):1209-16. doi: 10.1093/carcin/bgu099. Epub 2014 Apr 18. Carcinogenesis. 2014. PMID: 24747961 Free PMC article. Review. - A step-by-step microRNA guide to cancer development and metastasis.
Markopoulos GS, Roupakia E, Tokamani M, Chavdoula E, Hatziapostolou M, Polytarchou C, Marcu KB, Papavassiliou AG, Sandaltzopoulos R, Kolettas E. Markopoulos GS, et al. Cell Oncol (Dordr). 2017 Aug;40(4):303-339. doi: 10.1007/s13402-017-0341-9. Epub 2017 Jul 26. Cell Oncol (Dordr). 2017. PMID: 28748501 Review. - A component of the mir-17-92 polycistronic oncomir promotes oncogene-dependent apoptosis.
Olive V, Sabio E, Bennett MJ, De Jong CS, Biton A, McGann JC, Greaney SK, Sodir NM, Zhou AY, Balakrishnan A, Foth M, Luftig MA, Goga A, Speed TP, Xuan Z, Evan GI, Wan Y, Minella AC, He L. Olive V, et al. Elife. 2013 Oct 15;2:e00822. doi: 10.7554/eLife.00822. Elife. 2013. PMID: 24137534 Free PMC article.
Cited by
- RNA regulation of the immune system.
Ansel KM. Ansel KM. Immunol Rev. 2013 May;253(1):5-11. doi: 10.1111/imr.12062. Immunol Rev. 2013. PMID: 23550634 Free PMC article. No abstract available. - Limited miR-17-92 overexpression drives hematologic malignancies.
Danielson LS, Reavie L, Coussens M, Davalos V, Castillo-Martin M, Guijarro MV, Coffre M, Cordon-Cardo C, Aifantis I, Ibrahim S, Liu C, Koralov SB, Hernando E. Danielson LS, et al. Leuk Res. 2015 Mar;39(3):335-41. doi: 10.1016/j.leukres.2014.12.002. Epub 2014 Dec 10. Leuk Res. 2015. PMID: 25597017 Free PMC article. - MicroRNAs Regulating Tumor Immune Response in the Prediction of the Outcome in Patients With Breast Cancer.
Thomopoulou K, Papadaki C, Monastirioti A, Koronakis G, Mala A, Kalapanida D, Mavroudis D, Agelaki S. Thomopoulou K, et al. Front Mol Biosci. 2021 Jun 9;8:668534. doi: 10.3389/fmolb.2021.668534. eCollection 2021. Front Mol Biosci. 2021. PMID: 34179081 Free PMC article. - Hsa-miR-19a is associated with lymph metastasis and mediates the TNF-α induced epithelial-to-mesenchymal transition in colorectal cancer.
Huang L, Wang X, Wen C, Yang X, Song M, Chen J, Wang C, Zhang B, Wang L, Iwamoto A, Wang J, Liu H. Huang L, et al. Sci Rep. 2015 Aug 25;5:13350. doi: 10.1038/srep13350. Sci Rep. 2015. PMID: 26302825 Free PMC article. - The Nefarious Nexus of Noncoding RNAs in Cancer.
Anastasiadou E, Faggioni A, Trivedi P, Slack FJ. Anastasiadou E, et al. Int J Mol Sci. 2018 Jul 17;19(7):2072. doi: 10.3390/ijms19072072. Int J Mol Sci. 2018. PMID: 30018188 Free PMC article. Review.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. - PubMed
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science (New York, N.Y.) 2005;309:1519–24. - PubMed
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews. Molecular cell biology. 2009;10:126–39. - PubMed
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews. Genetics. 2008;9:102–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources