Cohabiting family members share microbiota with one another and with their dogs - PubMed (original) (raw)
Comparative Study
doi: 10.7554/eLife.00458.
Christian Lauber, Elizabeth K Costello, Catherine A Lozupone, Gregory Humphrey, Donna Berg-Lyons, J Gregory Caporaso, Dan Knights, Jose C Clemente, Sara Nakielny, Jeffrey I Gordon, Noah Fierer, Rob Knight
Affiliations
- PMID: 23599893
- PMCID: PMC3628085
- DOI: 10.7554/eLife.00458
Comparative Study
Cohabiting family members share microbiota with one another and with their dogs
Se Jin Song et al. Elife. 2013.
Abstract
Human-associated microbial communities vary across individuals: possible contributing factors include (genetic) relatedness, diet, and age. However, our surroundings, including individuals with whom we interact, also likely shape our microbial communities. To quantify this microbial exchange, we surveyed fecal, oral, and skin microbiota from 60 families (spousal units with children, dogs, both, or neither). Household members, particularly couples, shared more of their microbiota than individuals from different households, with stronger effects of co-habitation on skin than oral or fecal microbiota. Dog ownership significantly increased the shared skin microbiota in cohabiting adults, and dog-owning adults shared more 'skin' microbiota with their own dogs than with other dogs. Although the degree to which these shared microbes have a true niche on the human body, vs transient detection after direct contact, is unknown, these results suggest that direct and frequent contact with our cohabitants may significantly shape the composition of our microbial communities. DOI:http://dx.doi.org/10.7554/eLife.00458.001.
Keywords: Human; companion animals; environmental microbial reservoirs; family structure; metagenomics; microbial community transmission.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
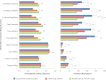
Figure 1.. Community similarity within and between families across body sites, and taxa contributing to these differences.
Panels (A–D) show average unweighted UniFrac distances between family members (blue) and between members of different families (red). ‘Child’ refers to all offspring aged 3–18 years who cohabit with the parents. ‘Infants’ were considered to be individuals aged 0–12 months. Palm/Paw refers to the right palm in the human comparisons and the back left paw in the dog comparison. Although there are distinguishable differences between the left and right palm communities within and across individuals (Fierer et al., 2008), the same analysis using the left palms showed a similar pattern (Table 2) and neither composition nor diversity were different enough between palms or among the four dog paws to affect the overall patterns. Mean ± 95% CI and R values (ANOSIM) are shown. *p<0.05 and **p<0.001 based on 10,000 permutations. Panel (E) shows the families of bacteria that exhibit the greatest differences in the number of phylotypes (OTUs) shared within and between adult partners on the right palm. Bars represent the average number of shared phylotypes for a given bacterial family within partners from the same family (blue) and between partners of different families (red). Mean ± 95% CI shown. *p<0.05 after Bonferroni correction (Wilcoxon test). DOI:
http://dx.doi.org/10.7554/eLife.00458.006
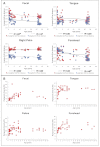
Figure 2.. Approach towards or departure from the ‘adult’ state in each body site with age.
(A) Each point represents the average distance (unweighted UniFrac in red; weighted UniFrac in blue) between each participant and all other participants in the ‘adult’ age bracket. Here we define baseline ‘adult’ as 30–45 years in age (the results are not sensitive to this threshold). R2 values (linear regression model) are shown. *p<0.01, **p<0.001. (B) Phylogenetic diversity (PD) of the communities on each body site is plotted for all of the offspring in the study (aged 0–18 years). DOI:
http://dx.doi.org/10.7554/eLife.00458.008
Figure 3.. Community similarity and phylotype sharing between dogs-owners and their dogs.
The left panel shows the average unweighted UniFrac distance between adult dog-owners and their dogs (blue), between dog-owners and other (not their own) dogs (red), and between adults who do not own dogs and dogs (green). The right panel shows the number of phylotypes shared for the same categories. Comparisons are labeled on the y-axis such that the first body site listed corresponds to the dog and the second site corresponds to the human. Mean ± 95% CI shown. The presence of asterisks lacking brackets indicates that all pairwise comparisons within that group are significant. Generally, dog-owners tend to share more similar communities and more phylotypes with their own dogs than with other dogs. *p<0.05, **p<0.001 after Bonferroni correction (Wilcoxon test). DOI:
http://dx.doi.org/10.7554/eLife.00458.011
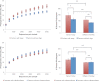
Figure 4.. Alpha diversity and shared phylotypes in couples with and without dogs and children.
The left panels show rarefaction curves for skin communities of couples (including seniors) who have dogs (top, in red), those without dogs (top, in blue), couples (excluding seniors) with infants/children (bottom, in red), and those without infants/children (bottom, in blue). Mean ± 95% CI shown. The right panels show the average number of phylotypes shared among individuals from the same categories shown in the left panels. Mean ± 95% CI shown. *p<0.05, **p<0.001 after Bonferroni correction (Wilcoxon test). DOI:
http://dx.doi.org/10.7554/eLife.00458.012
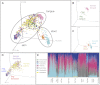
Figure 5.. Variation within and between the communities of skin, oral, and fecal samples from humans and dogs.
Panel (A) shows a PCoA plot of all the body habitats, using unweighted UniFrac distances of human and dog samples, rarefied at 5000 sequences/sample. Panels (B–D) show select body habitats from the full plot. Panel (E) shows a summary of the taxa shaded by relative abundance at the phylum level broken down by specific body habitat; the seven most abundant taxa are shown in the legend. DOI:
http://dx.doi.org/10.7554/eLife.00458.016
Similar articles
- Factors associated with dog ownership and contact with dogs in a UK community.
Westgarth C, Pinchbeck GL, Bradshaw JW, Dawson S, Gaskell RM, Christley RM. Westgarth C, et al. BMC Vet Res. 2007 Apr 3;3:5. doi: 10.1186/1746-6148-3-5. BMC Vet Res. 2007. PMID: 17407583 Free PMC article. - Interplay of personal, pet, and environmental colonization in households affected by community-associated methicillin-resistant Staphylococcus aureus.
Hogan PG, Mork RL, Boyle MG, Muenks CE, Morelli JJ, Thompson RM, Sullivan ML, Gehlert SJ, Merlo JR, McKenzie MG, Wardenburg JB, Rzhetsky A, Burnham CD, Fritz SA. Hogan PG, et al. J Infect. 2019 Mar;78(3):200-207. doi: 10.1016/j.jinf.2018.11.006. Epub 2018 Nov 29. J Infect. 2019. PMID: 30503843 Free PMC article. - Household Transmission of Clostridium difficile to Family Members and Domestic Pets.
Loo VG, Brassard P, Miller MA. Loo VG, et al. Infect Control Hosp Epidemiol. 2016 Nov;37(11):1342-1348. doi: 10.1017/ice.2016.178. Infect Control Hosp Epidemiol. 2016. PMID: 27767004 - Gut microbiota of humans, dogs and cats: current knowledge and future opportunities and challenges.
Deng P, Swanson KS. Deng P, et al. Br J Nutr. 2015 Jan;113 Suppl:S6-17. doi: 10.1017/S0007114514002943. Epub 2014 Nov 21. Br J Nutr. 2015. PMID: 25414978 Review. - Dogs' Microbiome From Tip to Toe.
Pereira AM, Clemente A. Pereira AM, et al. Top Companion Anim Med. 2021 Nov;45:100584. doi: 10.1016/j.tcam.2021.100584. Epub 2021 Sep 10. Top Companion Anim Med. 2021. PMID: 34509665 Review.
Cited by
- Changes in social environment impact primate gut microbiota composition.
Pearce CS, Bukovsky D, Douchant K, Katoch A, Greenlaw J, Gale DJ, Nashed JY, Brien D, Kuhlmeier VA, Sabbagh MA, Blohm G, De Felice FG, Pare M, Cook DJ, Scott SH, Munoz DP, Sjaarda CP, Tusche A, Sheth PM, Winterborn A, Boehnke S, Gallivan JP. Pearce CS, et al. Anim Microbiome. 2024 Nov 13;6(1):66. doi: 10.1186/s42523-024-00355-y. Anim Microbiome. 2024. PMID: 39538341 Free PMC article. - Microbiome transfer from native to invasive species may increase invasion risk.
Martignoni MM, Kolodny O. Martignoni MM, et al. Proc Biol Sci. 2024 Nov;291(2034):20241318. doi: 10.1098/rspb.2024.1318. Epub 2024 Nov 6. Proc Biol Sci. 2024. PMID: 39500380 Free PMC article. - The gut microbiota is essential for Trichinella spiralis-evoked suppression of colitis.
Sun H, Long SR, Jiang M, Zhang HR, Wang JJ, Liao ZX, Cui J, Wang ZQ. Sun H, et al. PLoS Negl Trop Dis. 2024 Nov 4;18(11):e0012645. doi: 10.1371/journal.pntd.0012645. eCollection 2024 Nov. PLoS Negl Trop Dis. 2024. PMID: 39495798 Free PMC article. - The human gut metacommunity as a conceptual aid in the development of precision medicine.
Tannock GW. Tannock GW. Front Microbiol. 2024 Oct 10;15:1469543. doi: 10.3389/fmicb.2024.1469543. eCollection 2024. Front Microbiol. 2024. PMID: 39464395 Free PMC article. - Comparative analysis based on shared amplicon sequence variants reveals that cohabitation influences gut microbiota sharing between humans and dogs.
Ito Y, Nagasawa M, Koyama K, Ito K, Kikusui T. Ito Y, et al. Front Vet Sci. 2024 Oct 7;11:1417461. doi: 10.3389/fvets.2024.1417461. eCollection 2024. Front Vet Sci. 2024. PMID: 39434718 Free PMC article.
References
- Anderson M, Gorley R, Clarke K. 2008. PERMANOVA+ for PRIMER: guide to software and statistical methods. PRIMER-E, Plymouth, UK
- Bates D, Maechler M, Dai B. 2008. The lme4 package. http://lme4.r-forge.r-project.org/
Publication types
MeSH terms
Grants and funding
- K01 DK090285/DK/NIDDK NIH HHS/United States
- R01 HG004872/HG/NHGRI NIH HHS/United States
- U01 HG004866/HG/NHGRI NIH HHS/United States
- HG4872, HG4866/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical