Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe - PubMed (original) (raw)
doi: 10.3201/eid1903.121503.
Heather J Baldwin, Victor Max Corman, Stefan M Klose, Michael Owusu, Evans Ewald Nkrumah, Ebenezer Kofi Badu, Priscilla Anti, Olivia Agbenyega, Benjamin Meyer, Samuel Oppong, Yaw Adu Sarkodie, Elisabeth K V Kalko, Peter H C Lina, Elena V Godlevska, Chantal Reusken, Antje Seebens, Florian Gloza-Rausch, Peter Vallo, Marco Tschapka, Christian Drosten, Jan Felix Drexler
Affiliations
- PMID: 23622767
- PMCID: PMC3647674
- DOI: 10.3201/eid1903.121503
Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe
Augustina Annan et al. Emerg Infect Dis. 2013 Mar.
Abstract
We screened fecal specimens of 4,758 bats from Ghana and 272 bats from 4 European countries for betacoronaviruses. Viruses related to the novel human betacoronavirus EMC/2012 were detected in 46 (24.9%) of 185 Nycteris bats and 40 (14.7%) of 272 Pipistrellus bats. Their genetic relatedness indicated EMC/2012 originated from bats.
Keywords: Africa; CoV-EMC; Europe; Ghana; bats; coronavirus; human betacoronavirus; viruses.
Figures
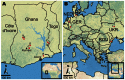
Figure 1
Location of bat sampling sites in Ghana and Europe. The 7 sites in Ghana (A) and the 5 areas in Europe (B) are marked with dots and numbered from west to east. a, Bouyem (N7°43′24.899′′ W1°59′16.501′′); b, Forikrom (N7°35′23.1′′ W1°52′30.299′′); c, Bobiri (N6°41′13.56′′ W1°20′38.94′′); d, Kwamang (N6°58′0.001′′ W1°16′0.001′′); e, Shai Hills (N5°55′44.4′′ E0°4′30′′); f, Akpafu Todzi (N7°15′43.099′′ E0°29′29.501′′); g, Likpe Todome (N7°9′50.198′′ E0°36′28.501′′); h, Province Gelderland, NED (N52°1′46.859′′ E6°13′4.908′′); i, Eifel area, federal state Rhineland-Palatinate, GER (N50°20′5.316′′ E7°14′30.912′′); j, Holstein area, federal state Schleswig-Holstein, GER (N54°14′51.271′′ E10°4′3.347′′); k, Tulcea county, ROU (N45°12′0.00′′ E29°0′0.00′′); l, Kiev region, UKR (N50°27′0.324′′ E30°31′24.24′′). NED, the Netherlands; GER, Germany; ROU, Romania; UKR, Ukraine.
Figure 2
RNA-dependent RNA polymerase (RdRp) gene and Spike genephylogenies including the novel betacoronaviruses from bats in Ghana and Europe. A) Bayesian phylogeny of an 816-nt RdRp gene sequence fragment corresponding to positions 14781–15596 in severe acute respiratory syndrome coronavirus (SARS-CoV) strain Frankfurt 1 (GenBank accession no. AY291315). Data were analyzed with MrBayes version 3.1 (
http://mrbayes.sourceforge.net/
) by using a WAG amino acid substitution model and 4 million generations sampled every 100 steps. Trees were annotated by using a burn-in of 10,000 and visualized with FigTree version 1.6.1 from the BEAST package (
). A whale gammacoronavirus was used as an outgroup. The novel Nycteris bat viruses are shown in boldface and red, the novel Pipistrellus bat viruses and other bat CoVs in the 2c clade are shown in boldface and cyan, and the novel human betacoronavirus EMC/2012 is shown in boldface. Values at deep nodes represent statistical support of grouping by posterior probabilities. CoV clades are depicted to the right of taxa. B) Phylogeny of the complete Spike gene of clade 2c CoVs determined by using the neighbor-joining method with an amino acid percentage distance substitution model and the complete deletion option in MEGA5 (
). The Nycteris CoV Spike gene was equidistant from other 2c-CoV Spike genes with 45.6%–46.8% aa divergence. Human coronavirus (hCoV)–OC43 was used as an outgroup. No complete Spike gene sequence was available for VM314 or the novel Pipistrellus bat CoVs. Scale bar represents percentage amino acid distance. The analysis comprised 1,731 aa residues. C) Phylogeny of the partial Spike gene of clade 2c CoVs, including the novel CoVs of Pipistrellus bats from Europe, determined by using a nucleotide distance substitution model and the complete deletion option in MEGA5. Scale bar represents percentage nucleotide distance. The analysis comprised 131 nt corresponding to positions 25378–25517 in hCoV-EMC/2012. Oligonucleotide sequences of primers used to amplify full and partial Spike gene sequences are available on request from the authors. Values at deep nodes in B and C represent statistical support of grouping by percentage of 1,000 bootstrap replicates. GenBank accession numbers for the complete and partial Spike genes correspond to those given in panel A for the RdRp gene.
Similar articles
- Alphacoronaviruses Detected in French Bats Are Phylogeographically Linked to Coronaviruses of European Bats.
Goffard A, Demanche C, Arthur L, Pinçon C, Michaux J, Dubuisson J. Goffard A, et al. Viruses. 2015 Dec 2;7(12):6279-90. doi: 10.3390/v7122937. Viruses. 2015. PMID: 26633467 Free PMC article. - First Report of Coronaviruses in Northern European Bats.
Kivistö I, Tidenberg EM, Lilley T, Suominen K, Forbes KM, Vapalahti O, Huovilainen A, Sironen T. Kivistö I, et al. Vector Borne Zoonotic Dis. 2020 Feb;20(2):155-158. doi: 10.1089/vbz.2018.2367. Epub 2019 Sep 10. Vector Borne Zoonotic Dis. 2020. PMID: 31503522 - [A novel coronavirus, MERS-CoV].
Mizutani T. Mizutani T. Uirusu. 2013;63(1):1-6. doi: 10.2222/jsv.63.1. Uirusu. 2013. PMID: 24769571 Review. Japanese. - Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic?
Chan JF, Li KS, To KK, Cheng VC, Chen H, Yuen KY. Chan JF, et al. J Infect. 2012 Dec;65(6):477-89. doi: 10.1016/j.jinf.2012.10.002. Epub 2012 Oct 13. J Infect. 2012. PMID: 23072791 Free PMC article. Review.
Cited by
- Wide Diversity of Coronaviruses in Frugivorous and Insectivorous Bat Species: A Pilot Study in Guinea, West Africa.
Lacroix A, Vidal N, Keita AK, Thaurignac G, Esteban A, De Nys H, Diallo R, Toure A, Goumou S, Soumah AK, Povogui M, Koivogui J, Monemou JL, Raulino R, Nkuba A, Foulongne V, Delaporte E, Ayouba A, Peeters M. Lacroix A, et al. Viruses. 2020 Aug 5;12(8):855. doi: 10.3390/v12080855. Viruses. 2020. PMID: 32764506 Free PMC article. - Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond.
Lu G, Wang Q, Gao GF. Lu G, et al. Trends Microbiol. 2015 Aug;23(8):468-78. doi: 10.1016/j.tim.2015.06.003. Epub 2015 Jul 21. Trends Microbiol. 2015. PMID: 26206723 Free PMC article. Review. - Design of Potential RNAi (miRNA and siRNA) Molecules for Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Gene Silencing by Computational Method.
Nur SM, Hasan MA, Amin MA, Hossain M, Sharmin T. Nur SM, et al. Interdiscip Sci. 2015 Sep;7(3):257-65. doi: 10.1007/s12539-015-0266-9. Epub 2015 Jul 30. Interdiscip Sci. 2015. PMID: 26223545 Free PMC article. - Coronavirus Spike Protein and Tropism Changes.
Hulswit RJ, de Haan CA, Bosch BJ. Hulswit RJ, et al. Adv Virus Res. 2016;96:29-57. doi: 10.1016/bs.aivir.2016.08.004. Epub 2016 Sep 13. Adv Virus Res. 2016. PMID: 27712627 Free PMC article. Review. - Bat-associated microbes: Opportunities and perils, an overview.
Dhivahar J, Parthasarathy A, Krishnan K, Kovi BS, Pandian GN. Dhivahar J, et al. Heliyon. 2023 Nov 18;9(12):e22351. doi: 10.1016/j.heliyon.2023.e22351. eCollection 2023 Dec. Heliyon. 2023. PMID: 38125540 Free PMC article. Review.
References
- Saif LJ. Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? Rev Sci Tech. 2004;23:643–60 . - PubMed
- Drexler JF, Gloza-Rausch F, Glende J, Corman VM, Muth D, Goettsche M, et al. Genomic characterization of severe acute respiratory syndrome–related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J Virol. 2010;84:11336–49. 10.1128/JVI.00650-10 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources